Discovery of Guanfacine as a Novel TAAR1 Agonist: A Combination Strategy through Molecular Modeling Studies and Biological Assays
- PMID: 38004497
- PMCID: PMC10674299
- DOI: 10.3390/ph16111632
Discovery of Guanfacine as a Novel TAAR1 Agonist: A Combination Strategy through Molecular Modeling Studies and Biological Assays
Abstract
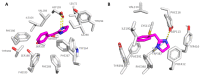
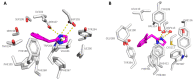
Trace amine-associated receptor 1 (TAAR1) is an attractive target for the design of innovative drugs to be applied in diverse pharmacological settings. Due to a non-negligible structural similarity with endogenous ligands, most of the agonists developed so far resulted in being affected by a low selectivity for TAAR1 with respect to other monoaminergic G protein-coupled receptors, like the adrenoreceptors. This study utilized comparative molecular docking studies and quantitative-structure activity relationship (QSAR) analyses to unveil key structural differences between TAAR1 and alpha2-adrenoreceptor (α2-ADR), with the aim to design novel TAAR1 agonists characterized by a higher selectivity profile and reduced off-target effects. While the presence of hydrophobic motives is encouraged towards both the two receptors, the introduction of polar/positively charged groups and the ligand conformation deeply affect the TAAR1 or α2-ADR putative selectivity. These computational methods allowed the identification of the α2A-ADR agonist guanfacine as an attractive TAAR1-targeting lead compound, demonstrating nanomolar activity in vitro. In vivo exploration of the efficacy of guanfacine showed that it is able to decrease the locomotor activity of dopamine transporter knockout (DAT-KO) rats. Therefore, guanfacine can be considered as an interesting template molecule worthy of structural optimization. The dual activity of guanfacine on both α2-ADR and TAAR1 signaling and the related crosstalk between the two pathways will deserve more in-depth investigation.
Keywords: AlphaFold; GPCR; QSAR; TAAR1; docking; dopamine; guanfacine; α2-adrenoreceptor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


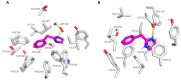









Similar articles
-
Unlocking the secrets of trace amine-associated receptor 1 agonists: new horizon in neuropsychiatric treatment.Front Psychiatry. 2024 Oct 31;15:1464550. doi: 10.3389/fpsyt.2024.1464550. eCollection 2024. Front Psychiatry. 2024. PMID: 39553890 Free PMC article. Review.
-
Computational Methods for the Discovery and Optimization of TAAR1 and TAAR5 Ligands.Int J Mol Sci. 2024 Jul 27;25(15):8226. doi: 10.3390/ijms25158226. Int J Mol Sci. 2024. PMID: 39125796 Free PMC article. Review.
-
Discovery of a Novel Chemo-Type for TAAR1 Agonism via Molecular Modeling.Molecules. 2024 Apr 11;29(8):1739. doi: 10.3390/molecules29081739. Molecules. 2024. PMID: 38675561 Free PMC article.
-
Normal thermoregulatory responses to 3-iodothyronamine, trace amines and amphetamine-like psychostimulants in trace amine associated receptor 1 knockout mice.J Neurosci Res. 2010 Jul;88(9):1962-9. doi: 10.1002/jnr.22367. J Neurosci Res. 2010. PMID: 20155805 Free PMC article.
-
The anticonvulsant and proconvulsant effects of alpha2-adrenoreceptor agonists are mediated by distinct populations of alpha2A-adrenoreceptors.Neuroscience. 2004;126(3):795-803. doi: 10.1016/j.neuroscience.2004.04.030. Neuroscience. 2004. PMID: 15183527
Cited by
-
Unlocking the secrets of trace amine-associated receptor 1 agonists: new horizon in neuropsychiatric treatment.Front Psychiatry. 2024 Oct 31;15:1464550. doi: 10.3389/fpsyt.2024.1464550. eCollection 2024. Front Psychiatry. 2024. PMID: 39553890 Free PMC article. Review.
-
Modulating autism spectrum disorder pathophysiology using a trace amine-focused approach: targeting the gut.Mol Med. 2025 May 20;31(1):198. doi: 10.1186/s10020-025-01232-3. Mol Med. 2025. PMID: 40394473 Free PMC article. Review.
-
Computational Methods for the Discovery and Optimization of TAAR1 and TAAR5 Ligands.Int J Mol Sci. 2024 Jul 27;25(15):8226. doi: 10.3390/ijms25158226. Int J Mol Sci. 2024. PMID: 39125796 Free PMC article. Review.
-
Discovery of a Novel Chemo-Type for TAAR1 Agonism via Molecular Modeling.Molecules. 2024 Apr 11;29(8):1739. doi: 10.3390/molecules29081739. Molecules. 2024. PMID: 38675561 Free PMC article.
References
-
- Zhai L., Xiao H., Lin C., Wong H.L.X., Lam Y.Y., Gong M., Wu G., Ning Z., Huang C., Zhang Y., et al. Gut Microbiota-Derived Tryptamine and Phenethylamine Impair Insulin Sensitivity in Metabolic Syndrome and Irritable Bowel Syndrome. Nat. Commun. 2023;14:4986. doi: 10.1038/s41467-023-40552-y. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

