Characterization of SARS-CoV-2 Variants in Military and Civilian Personnel of an Air Force Airport during Three Pandemic Waves in Italy
- PMID: 38004723
- PMCID: PMC10672769
- DOI: 10.3390/microorganisms11112711
Characterization of SARS-CoV-2 Variants in Military and Civilian Personnel of an Air Force Airport during Three Pandemic Waves in Italy
Abstract
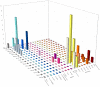
We investigated SARS-CoV-2 variants circulating, from November 2020 to March 2022, among military and civilian personnel at an Air Force airport in Italy in order to classify viral isolates in a potential hotspot for virus spread. Positive samples were subjected to Next-Generation Sequencing (NGS) of the whole viral genome and Sanger sequencing of the spike coding region. Phylogenetic analysis classified viral isolates and traced their evolutionary relationships. Clusters were identified using 70% cut-off. Sequencing methods yielded comparable results in terms of variant classification. In 2020 and 2021, we identified several variants, including B.1.258 (4/67), B.1.177 (9/67), Alpha (B.1.1.7, 9/67), Gamma (P.1.1, 4/67), and Delta (4/67). In 2022, only Omicron and its sub-lineage variants were observed (37/67). SARS-CoV-2 isolates were screened to detect naturally occurring resistance in genomic regions, the target of new therapies, comparing them to the Wuhan Hu-1 reference strain. Interestingly, 2/30 non-Omicron isolates carried the G15S 3CLpro substitution responsible for reduced susceptibility to protease inhibitors. On the other hand, Omicron isolates carried unusual substitutions A1803V, D1809N, and A949T on PLpro, and the D216N on 3CLpro. Finally, the P323L substitution on RdRp coding regions was not associated with the mutational pattern related to polymerase inhibitor resistance. This study highlights the importance of continuous genomic surveillance to monitor SARS-CoV-2 evolution in the general population, as well as in restricted communities.
Keywords: 3CLpro; Next-Generation Sequencing; Plpro; RdRp; SARS-CoV-2 variant; Sanger; mutations; nucleocapsid; spike.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Receptor-Binding-Motif-Targeted Sanger Sequencing: a Quick and Cost-Effective Strategy for Molecular Surveillance of SARS-CoV-2 Variants.Microbiol Spectr. 2022 Jun 29;10(3):e0066522. doi: 10.1128/spectrum.00665-22. Epub 2022 May 31. Microbiol Spectr. 2022. PMID: 35638906 Free PMC article.
-
A tool for the cheap and rapid screening of SARS-CoV-2 variants of concern (VoCs) by Sanger sequencing.Microbiol Spectr. 2023 Sep 7;11(5):e0506422. doi: 10.1128/spectrum.05064-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 37676038 Free PMC article.
-
Two-Period Study Results from a Large Italian Hospital Laboratory Attesting SARS-CoV-2 Variant PCR Assay Evolution.Microbiol Spectr. 2022 Dec 21;10(6):e0292222. doi: 10.1128/spectrum.02922-22. Epub 2022 Nov 21. Microbiol Spectr. 2022. PMID: 36409091 Free PMC article.
-
Molecular evolution of SARS-CoV-2 from December 2019 to August 2022.J Med Virol. 2023 Jan;95(1):e28366. doi: 10.1002/jmv.28366. J Med Virol. 2023. PMID: 36458547 Free PMC article. Review.
-
The Rise and Fall of SARS-CoV-2 Variants and Ongoing Diversification of Omicron.Viruses. 2022 Sep 10;14(9):2009. doi: 10.3390/v14092009. Viruses. 2022. PMID: 36146815 Free PMC article. Review.
Cited by
-
Wastewater sequencing from a rural community enables identification of widespread adaptive mutations in a SARS-CoV-2 alpha variant.Sci Rep. 2025 May 28;15(1):18657. doi: 10.1038/s41598-025-03771-5. Sci Rep. 2025. PMID: 40437225 Free PMC article.
-
Surfing the Waves of SARS-CoV-2: Analysis of Viral Genome Variants Using an NGS Survey in Verona, Italy.Microorganisms. 2024 Apr 24;12(5):846. doi: 10.3390/microorganisms12050846. Microorganisms. 2024. PMID: 38792676 Free PMC article.
-
Prevalence of Enteric Pathogens and Antibiotic Resistance: Results of a Six-Year Active Surveillance Study on Patients Admitted to a Teaching Hospital.Antibiotics (Basel). 2024 Aug 2;13(8):726. doi: 10.3390/antibiotics13080726. Antibiotics (Basel). 2024. PMID: 39200026 Free PMC article.
References
-
- Fabiani M., Ramigni M., Gobbetto V., Mateo-Urdiales A., Pezzotti P., Piovesan C. Effectiveness of the Comirnaty (BNT162b2, BioNTech/Pfizer) vaccine in preventing SARS-CoV-2 infection among healthcare workers, Treviso province, Veneto region, Italy, 27 December 2020 to 24 March 2021. Euro Surveill. 2021;26:2100420. doi: 10.2807/1560-7917.ES.2021.26.17.2100420. - DOI - PMC - PubMed
-
- Oliani F., Savoia A., Gallo G., Tiwana N., Letzgus M., Gentiloni F., Piatti A., Chiappa L., Bisesti A., Laquintana D., et al. Italy’s rollout of COVID-19 vaccinations: The crucial contribution of the first experimental mass vaccination site in Lombardy. Vaccine. 2022;40:1397–1403. doi: 10.1016/j.vaccine.2022.01.059. - DOI - PMC - PubMed
-
- Dhanasooraj D., Viswanathan P., Saphia S., Jose B.P., Parambath F.C., Sivadas S., Akash N.P., Vimisha T.V., Nair P.R., Mohan A., et al. Genomic surveillance of SARS-CoV-2 by sequencing the RBD region using Sanger sequencing from North Kerala. Front. Public Health. 2022;10:974667. doi: 10.3389/fpubh.2022.974667. - DOI - PMC - PubMed
-
- Guthrie J.L., Teatero S., Zittermann S., Chen Y., Sullivan A., Rilkoff H., Joshi E., Sivaraman K., de Borja R., Sundaravadanam Y., et al. Detection of the novel SARS-CoV-2 European lineage B.1.177 in Ontario, Canada. J. Clin. Virol. Plus. 2021;1:100010. doi: 10.1016/j.jcvp.2021.100010. - DOI - PMC - PubMed
-
- European Centre for Disease Prevention and Control ECDC. [(accessed on 11 September 2023)]. Available online: https://www.ecdc.europa.eu/en.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

