Resistance and Virulence Surveillance in Escherichia coli Isolated from Commercial Meat Samples: A One Health Approach
- PMID: 38004724
- PMCID: PMC10672981
- DOI: 10.3390/microorganisms11112712
Resistance and Virulence Surveillance in Escherichia coli Isolated from Commercial Meat Samples: A One Health Approach
Abstract
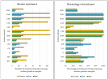
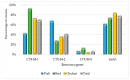
Escherichia coli is a key indicator of food hygiene, and its monitoring in meat samples points to the potential presence of antimicrobial-resistant strains capable of causing infections in humans, encompassing resistance profiles categorized as serious threats by the Centers for Disease Control and Prevention (CDC), such as Extended-Spectrum Beta-Lactamase (ESBL)-a problem with consequences for animal, human, and environmental health. The objective of the present work was to isolate and characterize ESBL-producing E. coli strains from poultry, pork, and beef meat samples, with a characterization of their virulence and antimicrobial resistance profiles. A total of 450 meat samples (150 chicken, 150 beef, and 150 pork) were obtained from supermarkets and subsequently cultured in medium supplemented with cefotaxime. The isolated colonies were characterized biochemically, followed by antibiogram testing using the disk diffusion technique. Further classification involved biofilm formation and the presence of antimicrobial resistance genes (blaCTX-M, AmpC-type, mcr-1, and fosA3), and virulence genes (eaeA, st, bfpA, lt, stx1, stx2, aggR, iss, ompT, hlyF, iutA, iroN, fyuA, cvaC, and hylA). Statistical analysis was performed via the likelihood-ratio test. In total, 168 strains were obtained, with 73% originating from chicken, 22% from pork, and 17% from beef samples. Notably, strains exhibited greater resistance to tetracycline (51%), ciprofloxacin (46%), and fosfomycin (38%), apart from β-lactams. The detection of antimicrobial resistance in food-isolated strains is noteworthy, underscoring the significance of antimicrobial resistance as a global concern. More than 90% of the strains were biofilm producers, and strains carrying many ExPEC genes were more likely to be biofilm formers (OR 2.42), which increases the problem since the microorganisms have a greater chance of environment persistence and genetic exchange. Regarding molecular characterization, bovine samples showed a higher prevalence of blaCTX-M-1 (OR 6.52), while chicken strains were more likely to carry the fosA3 gene (OR 2.43, CI 1.17-5.05) and presented between 6 to 8 ExPEC genes (OR 2.5, CI 1.33-5.01) compared to other meat samples. Concerning diarrheagenic E. coli genes, two strains harbored eae. It is important to highlight these strains, as they exhibited both biofilm-forming capacities and multidrug resistance (MDR), potentially enabling colonization in diverse environments and causing infections. In conclusion, this study underscores the presence of β-lactamase-producing E. coli strains, mainly in poultry samples, compared to beef and pork samples. Furthermore, all meat sample strains exhibited many virulence-associated extraintestinal genes, with some strains harboring diarrheagenic E. coli (DEC) genes.
Keywords: ESBL; biofilm formation; diarrheagenic Escherichia coli; extraintestinal Escherichia coli; meat samples; virulence genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Evaluation of the Antibiotic Resistance and Virulence of Escherichia coli Strains Isolated from Chicken Carcasses in 2007 and 2013 from Paraná, Brazil.Foodborne Pathog Dis. 2015 Jun;12(6):479-85. doi: 10.1089/fpd.2014.1888. Epub 2015 May 14. Foodborne Pathog Dis. 2015. PMID: 25974222
-
Occurrence of the Colistin Resistance Gene mcr-1 and Additional Antibiotic Resistance Genes in ESBL/AmpC-Producing Escherichia coli from Poultry in Lebanon: A Nationwide Survey.Microbiol Spectr. 2021 Oct 31;9(2):e0002521. doi: 10.1128/Spectrum.00025-21. Epub 2021 Sep 8. Microbiol Spectr. 2021. PMID: 34494875 Free PMC article.
-
Molecular detection of Shiga toxin and extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli isolates from sheep and goats.Mol Biol Rep. 2024 Jan 2;51(1):57. doi: 10.1007/s11033-023-08987-0. Mol Biol Rep. 2024. PMID: 38165462 Free PMC article.
-
Antimicrobial Resistance in Diverse Escherichia coli Pathotypes from Nigeria.Antibiotics (Basel). 2024 Sep 26;13(10):922. doi: 10.3390/antibiotics13100922. Antibiotics (Basel). 2024. PMID: 39452189 Free PMC article. Review.
-
Antimicrobial resistance in food-associated Escherichia coli in Mexico and Latin America.Biosci Microbiota Food Health. 2024;43(1):4-12. doi: 10.12938/bmfh.2023-022. Epub 2023 Jun 29. Biosci Microbiota Food Health. 2024. PMID: 38188662 Free PMC article. Review.
Cited by
-
Xenobiotics and Broiler Microbiota: Molecular Insights into Bacterial Antimicrobial Resistance and Food Safety Implications for Human Health.J Xenobiot. 2025 Aug 8;15(4):129. doi: 10.3390/jox15040129. J Xenobiot. 2025. PMID: 40863336 Free PMC article. Review.
-
Overview of Antimicrobial Resistant ESKAPEE Pathogens in Food Sources and Their Implications from a One Health Perspective.Microorganisms. 2024 Oct 18;12(10):2084. doi: 10.3390/microorganisms12102084. Microorganisms. 2024. PMID: 39458393 Free PMC article. Review.
References
-
- Cyoia P.S., Koga V.L., Nishio E.K., Houle S., Dozois C.M., de Brito K.C.T., de Brito B.G., Nakazato G., Kobayashi R.K.T. Distribution of ExPEC Virulence Factors, blaCTX-M, fosA3, and mcr-1 in Escherichia coli Isolated from Commercialized Chicken Carcasses. Front. Microbiol. 2019;9:3254. doi: 10.3389/fmicb.2018.03254. - DOI - PMC - PubMed
-
- Sharma J., Kumar D., Hussain S., Pathak A., Shukla M., Prasanna Kumar V., Anisha P.N., Rautela R., Upadhyay A.K., Singh S.P. Prevalence, Antimicrobial Resistance and Virulence Genes Characterization of Nontyphoidal Salmonella Isolated from Retail Chicken Meat Shops in Northern India. Food Control. 2019;102:104–111. doi: 10.1016/j.foodcont.2019.01.021. - DOI
LinkOut - more resources
Full Text Sources
Research Materials

