Genomic Island-Encoded Diguanylate Cyclase from Vibrio alginolyticus Regulates Biofilm Formation and Motility in Pseudoalteromonas
- PMID: 38004737
- PMCID: PMC10672970
- DOI: 10.3390/microorganisms11112725
Genomic Island-Encoded Diguanylate Cyclase from Vibrio alginolyticus Regulates Biofilm Formation and Motility in Pseudoalteromonas
Abstract
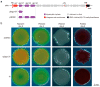
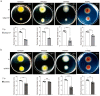
Many bacteria use the second messenger c-di-GMP to regulate exopolysaccharide production, biofilm formation, motility, virulence, and other phenotypes. The c-di-GMP level is controlled by the complex network of diguanylate cyclases (DGCs) and phosphodiesterases (PDEs) that synthesize and degrade c-di-GMP. In addition to chromosomally encoded DGCs, increasing numbers of DGCs were found to be located on mobile genetic elements. Whether these mobile genetic element-encoded DGCs can modulate the physiological phenotypes in recipient bacteria after horizontal gene transfer should be investigated. In our previous study, a genomic island encoding three DGC proteins (Dgc137, Dgc139, and Dgc140) was characterized in Vibrio alginolyticus isolated from the gastric cavity of the coral Galaxea fascicularis. Here, the effect of the three DGCs in four Pseudoalteromonas strains isolated from coral Galaxea fascicularis and other marine environments was explored. The results showed that when dgc137 is present rather than the three DGC genes, it obviously modulates biofilm formation and bacterial motility in these Pseudoalteromonas strains. Our findings implied that mobile genetic element-encoded DGC could regulate the physiological status of neighboring bacteria in a microbial community by modulating the c-di-GMP level after horizontal gene transfer.
Keywords: Pseudoalteromonas; Vibrio; biofilm; mobile genetic elements; motility.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Enzymatically active and inactive phosphodiesterases and diguanylate cyclases are involved in regulation of Motility or sessility in Escherichia coli CFT073.mBio. 2012 Oct 9;3(5):e00307-12. doi: 10.1128/mBio.00307-12. mBio. 2012. PMID: 23047748 Free PMC article.
-
More than Enzymes That Make or Break Cyclic Di-GMP-Local Signaling in the Interactome of GGDEF/EAL Domain Proteins of Escherichia coli.mBio. 2017 Oct 10;8(5):e01639-17. doi: 10.1128/mBio.01639-17. mBio. 2017. PMID: 29018125 Free PMC article.
-
The Inhibitory Site of a Diguanylate Cyclase Is a Necessary Element for Interaction and Signaling with an Effector Protein.J Bacteriol. 2016 May 13;198(11):1595-603. doi: 10.1128/JB.00090-16. Print 2016 Jun 1. J Bacteriol. 2016. PMID: 27002135 Free PMC article.
-
Screening for Diguanylate Cyclase (DGC) Inhibitors Mitigating Bacterial Biofilm Formation.Front Chem. 2020 Apr 21;8:264. doi: 10.3389/fchem.2020.00264. eCollection 2020. Front Chem. 2020. PMID: 32373581 Free PMC article. Review.
-
Diguanylate Cyclases in Vibrio cholerae: Essential Regulators of Lifestyle Switching.Front Cell Infect Microbiol. 2020 Oct 22;10:582947. doi: 10.3389/fcimb.2020.582947. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33194821 Free PMC article. Review.
Cited by
-
Extracellular Vesicles From Xylella fastidiosa Carry sRNAs and Genomic Islands, Suggesting Roles in Recipient Cells.J Extracell Vesicles. 2025 Jun;14(6):e70102. doi: 10.1002/jev2.70102. J Extracell Vesicles. 2025. PMID: 40560800 Free PMC article.
References
Grants and funding
- 42188102, 91951203 and 32070175/the National Science Foundation of China
- 2022YFC3103600/the National Key R&D Program of China
- 2021345/the Youth Innovation Promotion Association CAS
- 2019BT02Y262/the Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program
- GJTD-2020-12/the K. C. Wong Education Foundation
LinkOut - more resources
Full Text Sources

