Two-Dose Ceftiofur Treatment Increases Cephamycinase Gene Quantities and Fecal Microbiome Diversity in Dairy Cows Diagnosed with Metritis
- PMID: 38004740
- PMCID: PMC10673576
- DOI: 10.3390/microorganisms11112728
Two-Dose Ceftiofur Treatment Increases Cephamycinase Gene Quantities and Fecal Microbiome Diversity in Dairy Cows Diagnosed with Metritis
Abstract
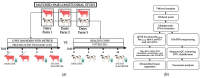
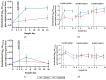
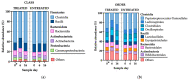
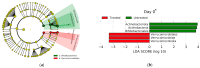
Antimicrobial resistance is a significant concern worldwide; meanwhile, the impact of 3rd generation cephalosporin (3GC) antibiotics on the microbial communities of cattle and resistance within these communities is largely unknown. The objectives of this study were to determine the effects of two-dose ceftiofur crystalline-free acid (2-CCFA) treatment on the fecal microbiota and on the quantities of second-and third-generation cephalosporin, fluoroquinolone, and macrolide resistance genes in Holstein-Friesian dairy cows in the southwestern United States. Across three dairy farms, 124 matched pairs of cows were enrolled in a longitudinal study. Following the product label regimen, CCFA was administered on days 0 and 3 to cows diagnosed with postpartum metritis. Healthy cows were pair-matched based on lactation number and calving date. Fecal samples were collected on days 0, 6, and 16 and pooled in groups of 4 (n = 192) by farm, day, and treatment group for community DNA extraction. The characterization of community DNA included real-time PCR (qPCR) to quantify the following antibiotic resistance genes: blaCMY-2, blaCTX-M, mphA, qnrB19, and the highly conserved 16S rRNA back-calculated to gene copies per gram of feces. Additionally, 16S rRNA amplicon sequencing and metagenomics analyses were used to determine differences in bacterial community composition by treatment, day, and farm. Overall, blaCMY-2 gene copies per gram of feces increased significantly (p ≤ 0.05) in the treated group compared to the untreated group on day 6 and remained elevated on day 16. However, blaCTX-M, mphA, and qnrB19 gene quantities did not differ significantly (p ≥ 0.05) between treatment groups, days, or farms, suggesting a cephamycinase-specific enhancement in cows on these farms. Perhaps unexpectedly, 16S rRNA amplicon metagenomic analyses showed that the fecal bacterial communities from treated animals on day 6 had significantly greater (p ≤ 0.05) alpha and beta diversity than the untreated group. Two-dose ceftiofur treatment in dairy cows with metritis elevates cephamycinase gene quantities among all fecal bacteria while paradoxically increasing microbial diversity.
Keywords: 3rd generation cephalosporin; antibiotic resistance genes; dairy cattle; ecological diversity; fecal microbiota; fluoroquinolone; macrolide; metagenomics; metritis; two-dose CCFA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Fleming A. Classics in infectious diseases: On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae by Alexander Fleming. Br. J. Exp. Pathol. 1929;10:226–236. reprinted in Rev. Infect. Dis. 1980, 2, 129–139. - PubMed
-
- CDC . Antibiotic Resistance Threats in the United States. U.S. Department of Health and Human Services; Atlanta, GA, USA: 2019.
-
- O’Neil J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Volume 1. Government of the United Kingdom; London, UK: 2016. p. 84.
Grants and funding
LinkOut - more resources
Full Text Sources

