Transcriptome and Secretome Analyses of Endophyte Methylobacterium mesophilicum and Pathogen Xylella fastidiosa Interacting Show Nutrient Competition
- PMID: 38004766
- PMCID: PMC10673610
- DOI: 10.3390/microorganisms11112755
Transcriptome and Secretome Analyses of Endophyte Methylobacterium mesophilicum and Pathogen Xylella fastidiosa Interacting Show Nutrient Competition
Abstract
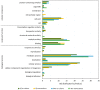
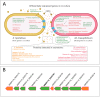
Xylella fastidiosa is the causal agent of several plant diseases affecting fruit and nut crops. Methylobacterium mesophilicum strain SR1.6/6 was isolated from Citrus sinensis and shown to promote plant growth by producing phytohormones, providing nutrients, inhibiting X. fastidiosa, and preventing Citrus Variegated Chlorosis. However, the molecular mechanisms involved in the interaction among these microbes are still unclear. The present work aimed to analyze physiological and molecular aspects of M. mesophilicum SR1.6/6 and X. fastidiosa 9a5c in co-culture. The transcriptome and secretome analyses indicated that X. fastidiosa down-regulates cell division and transport genes and up-regulates stress via induction of chaperones and pathogenicity-related genes including, the lipase-esterase LesA, a protease, as well as an oligopeptidase in response to M. mesophilicum competition. On the other hand, M. mesophilicum also down-regulated transport genes, except for iron uptake, which was up-regulated. Secretome analysis identified four proteins in M. mesophilicum exclusively produced in co-culture with X. fastidiosa, among these, three are related to phosphorous uptake. These results suggest that M. mesophilicum inhibits X. fastidiosa growth mainly due to nutrient competition for iron and phosphorous, thus promoting X. fastidiosa starvation, besides producing enzymes that degrade X. fastidiosa cell wall, mainly hydrolases. The understanding of these interactions provides a direction for control and management of the phytopathogen X. fastidiosa, and consequently, helps to improve citrus growth and productivity.
Keywords: citrus; endophytic bacteria; phytopathogen–endophyte interaction; secretome; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Differential gene expression in Xylella fastidiosa 9a5c during co-cultivation with the endophytic bacterium Methylobacterium mesophilicum SR1.6/6.J Basic Microbiol. 2015 Dec;55(12):1357-66. doi: 10.1002/jobm.201400916. Epub 2015 Jul 27. J Basic Microbiol. 2015. PMID: 26218710
-
Model plants for studying the interaction between Methylobacterium mesophilicum and Xylella fastidiosa.Can J Microbiol. 2006 May;52(5):419-26. doi: 10.1139/w05-142. Can J Microbiol. 2006. PMID: 16699566
-
Transmission of Methylobacterium mesophilicum by Bucephalogonia xanthophis for paratransgenic control strategy of citrus variegated chlorosis.J Microbiol. 2009 Aug;47(4):448-54. doi: 10.1007/s12275-008-0303-z. Epub 2009 Sep 9. J Microbiol. 2009. PMID: 19763419
-
A genomic approach to the understanding of Xylella fastidiosa pathogenicity.Curr Opin Microbiol. 2000 Oct;3(5):459-62. doi: 10.1016/s1369-5274(00)00121-1. Curr Opin Microbiol. 2000. PMID: 11050442 Review.
-
Etiology of three recent diseases of citrus in São Paulo State: sudden death, variegated chlorosis and huanglongbing.IUBMB Life. 2007 Apr-May;59(4-5):346-54. doi: 10.1080/15216540701299326. IUBMB Life. 2007. PMID: 17505974 Review.
Cited by
-
Diffusible Signal Factors and Xylella fastidiosa: A Crucial Mechanism Yet to Be Revealed.Biology (Basel). 2025 Mar 17;14(3):303. doi: 10.3390/biology14030303. Biology (Basel). 2025. PMID: 40136559 Free PMC article. Review.
-
The Role of Heat Shock Protein (Hsp) Chaperones in Environmental Stress Adaptation and Virulence of Plant Pathogenic Bacteria.Int J Mol Sci. 2025 Jan 9;26(2):528. doi: 10.3390/ijms26020528. Int J Mol Sci. 2025. PMID: 39859244 Free PMC article. Review.
References
-
- Wells J.M., Raju B.C., Hung H.-Y., Weisburg W.G., Mandelco-Paul L., Brenner D.J. Xylella fastidiosa gen. nov., sp. nov: Gram-negative, xylem-limited, fastidious plant bacteria related to Xanthomonas spp. Int. J. Syst. Bacteriol. 1987;37:136–143. doi: 10.1099/00207713-37-2-136. - DOI
-
- Hopkins D.L. Xylella fastidiosa: Xylem-limited bacterial pathogen of plants. Ann. Rev. Phytopathol. 1989;27:271–290. doi: 10.1146/annurev.py.27.090189.001415. - DOI
-
- Cariddi C., Saponari M., Boscia D., De Stradis A., Loconsole G., Nigro F., Porcelli F., Potere O., Martelli G.P. Isolation of a Xylella fastidiosa strain infecting olive and oleander in Apulia, Italy. J. Plant Pathol. 2014;96:425–429.

