The Microevolution of Antifungal Drug Resistance in Pathogenic Fungi
- PMID: 38004768
- PMCID: PMC10673521
- DOI: 10.3390/microorganisms11112757
The Microevolution of Antifungal Drug Resistance in Pathogenic Fungi
Abstract
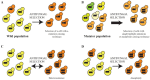
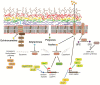
The mortality rates of invasive fungal infections remain high because of the limited number of antifungal drugs available and antifungal drug resistance, which can rapidly evolve during treatment. Mutations in key resistance genes such as ERG11 were postulated to be the predominant cause of antifungal drug resistance in the clinic. However, recent advances in whole genome sequencing have revealed that there are multiple mechanisms leading to the microevolution of resistance. In many fungal species, resistance can emerge through ERG11-independent mechanisms and through the accumulation of mutations in many genes to generate a polygenic resistance phenotype. In addition, genome sequencing has revealed that full or partial aneuploidy commonly occurs in clinical or microevolved in vitro isolates to confer antifungal resistance. This review will provide an overview of the mutations known to be selected during the adaptive microevolution of antifungal drug resistance and focus on how recent advances in genome sequencing technology have enhanced our understanding of this process.
Keywords: adaptation; aneuploidy; antifungal drugs; fungi; microevolution; mutator; pathogen; phenotypic diversity; ploidy; resistance.
Conflict of interest statement
The author declares no conflict of interest.
Figures
Similar articles
-
Mismatch Repair of DNA Replication Errors Contributes to Microevolution in the Pathogenic Fungus Cryptococcus neoformans.mBio. 2017 May 30;8(3):e00595-17. doi: 10.1128/mBio.00595-17. mBio. 2017. PMID: 28559486 Free PMC article.
-
Molecular mechanisms underlying the emergence of polygenetic antifungal drug resistance in msh2 mismatch repair mutants of Cryptococcus.JAC Antimicrob Resist. 2022 Apr 7;4(2):dlac033. doi: 10.1093/jacamr/dlac033. eCollection 2022 Apr. JAC Antimicrob Resist. 2022. PMID: 35402912 Free PMC article.
-
Genomic Diversity across Candida auris Clinical Isolates Shapes Rapid Development of Antifungal Resistance In Vitro and In Vivo.mBio. 2022 Aug 30;13(4):e0084222. doi: 10.1128/mbio.00842-22. Epub 2022 Jul 5. mBio. 2022. PMID: 35862787 Free PMC article.
-
Resistance in human pathogenic yeasts and filamentous fungi: prevalence, underlying molecular mechanisms and link to the use of antifungals in humans and the environment.Dan Med J. 2016 Oct;63(10):B5288. Dan Med J. 2016. PMID: 27697142 Review.
-
Ploidy dynamics and evolvability in fungi.Philos Trans R Soc Lond B Biol Sci. 2016 Dec 5;371(1709):20150461. doi: 10.1098/rstb.2015.0461. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 28080987 Free PMC article. Review.
Cited by
-
Molecular Mechanisms of Drug Resistance in Leishmania spp.Pathogens. 2024 Sep 27;13(10):835. doi: 10.3390/pathogens13100835. Pathogens. 2024. PMID: 39452707 Free PMC article. Review.
-
Evolution of the pathogenic mold Aspergillus fumigatus on high copper levels identifies novel resistance genes.mSphere. 2024 Jun 25;9(6):e0025324. doi: 10.1128/msphere.00253-24. Epub 2024 May 30. mSphere. 2024. PMID: 38814077 Free PMC article.
-
Resistance Mechanisms of Plant Pathogenic Fungi to Fungicide, Environmental Impacts of Fungicides, and Sustainable Solutions.Plants (Basel). 2024 Sep 30;13(19):2737. doi: 10.3390/plants13192737. Plants (Basel). 2024. PMID: 39409607 Free PMC article. Review.
-
Comparison and Analysis of Resistance Differences in Alternaria alternata from Fungicides with Three Different Mechanisms.J Fungi (Basel). 2025 Apr 11;11(4):305. doi: 10.3390/jof11040305. J Fungi (Basel). 2025. PMID: 40278125 Free PMC article.
-
Antifungal activity of guanidine compounds.Braz J Microbiol. 2025 Jun;56(2):1049-1059. doi: 10.1007/s42770-025-01625-w. Epub 2025 Feb 12. Braz J Microbiol. 2025. PMID: 39934527
References
Publication types
LinkOut - more resources
Full Text Sources

