Potent Antiviral Activity of Vitamin B12 against Severe Acute Respiratory Syndrome Coronavirus 2, Middle East Respiratory Syndrome Coronavirus, and Human Coronavirus 229E
- PMID: 38004788
- PMCID: PMC10673013
- DOI: 10.3390/microorganisms11112777
Potent Antiviral Activity of Vitamin B12 against Severe Acute Respiratory Syndrome Coronavirus 2, Middle East Respiratory Syndrome Coronavirus, and Human Coronavirus 229E
Abstract
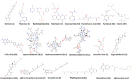
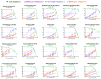
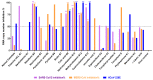
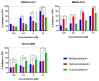
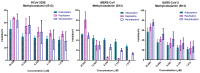
Repurposing vitamins as antiviral supporting agents is a rapid approach used to control emerging viral infections. Although there is considerable evidence supporting the use of vitamin supplementation in viral infections, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the specific role of each vitamin in defending against coronaviruses remains unclear. Antiviral activities of available vitamins on the infectivity and replication of human coronaviruses, namely, SARS-CoV-2, Middle East respiratory syndrome coronavirus (MERS-CoV), and human coronavirus 229E (HCoV-229E), were investigated using in silico and in vitro studies. We identified potential broad-spectrum inhibitor effects of Hydroxocobalamin and Methylcobalamin against the three tested CoVs. Cyanocobalamin could selectively affect SARS-CoV-2 but not MERS-CoV and HCoV-229E. Methylcobalamin showed significantly higher inhibition values on SARS-CoV-2 compared with Hydroxocobalamin and Cyanocobalamin, while Hydroxocobalamin showed the highest potent antiviral activity against MERS-CoV and Cyanocobalamin against HCoV-229E. Furthermore, in silico studies were performed for these promising vitamins to investigate their interaction with SARS-CoV-2, MERS-CoV, and HCoV-229E viral-specific cell receptors (ACE2, DPP4, and hAPN protein, respectively) and viral proteins (S-RBD, 3CL pro, RdRp), suggesting that Hydroxocobalamin, Methylcobalamin, and Cyanocobalamin may have significant binding affinity to these proteins. These results show that Methylcobalamin may have potential benefits for coronavirus-infected patients.
Keywords: MERS-CoV; SARS-CoV-2; antiviral agent; viral infection; vitamin B12; vitamins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
In vitro virucidal activity of Echinaforce®, an Echinacea purpurea preparation, against coronaviruses, including common cold coronavirus 229E and SARS-CoV-2.Virol J. 2020 Sep 9;17(1):136. doi: 10.1186/s12985-020-01401-2. Virol J. 2020. PMID: 32907596 Free PMC article.
-
Current Strategies of Antiviral Drug Discovery for COVID-19.Front Mol Biosci. 2021 May 13;8:671263. doi: 10.3389/fmolb.2021.671263. eCollection 2021. Front Mol Biosci. 2021. PMID: 34055887 Free PMC article. Review.
-
HTCC as a Polymeric Inhibitor of SARS-CoV-2 and MERS-CoV.J Virol. 2021 Jan 28;95(4):e01622-20. doi: 10.1128/JVI.01622-20. Print 2021 Jan 28. J Virol. 2021. PMID: 33219167 Free PMC article.
-
Transcriptomic profiling and genomic mutational analysis of Human coronavirus (HCoV)-229E -infected human cells.PLoS One. 2021 Feb 25;16(2):e0247128. doi: 10.1371/journal.pone.0247128. eCollection 2021. PLoS One. 2021. PMID: 33630927 Free PMC article.
-
An overview on the seven pathogenic human coronaviruses.Rev Med Virol. 2022 Mar;32(2):e2282. doi: 10.1002/rmv.2282. Epub 2021 Aug 2. Rev Med Virol. 2022. PMID: 34339073 Review.
Cited by
-
Immune-Boosting and Antiviral Effects of Antioxidants in COVID-19 Pneumonia: A Therapeutic Perspective.Life (Basel). 2025 Jan 16;15(1):113. doi: 10.3390/life15010113. Life (Basel). 2025. PMID: 39860053 Free PMC article. Review.
References
-
- Gaunt E.R., Hardie A., Claas E.C., Simmonds P., Templeton K.E. Epidemiology and Clinical Presentations of the Four Human Coronaviruses 229E, HKU1, NL63, and OC43 Detected over 3 Years Using a Novel Multiplex Real-Time PCR Method. J. Clin. Microbiol. 2010;48:2940–2947. doi: 10.1128/JCM.00636-10. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

