Methanomethylophilus alvi gen. nov., sp. nov., a Novel Hydrogenotrophic Methyl-Reducing Methanogenic Archaea of the Order Methanomassiliicoccales Isolated from the Human Gut and Proposal of the Novel Family Methanomethylophilaceae fam. nov
- PMID: 38004804
- PMCID: PMC10673518
- DOI: 10.3390/microorganisms11112794
Methanomethylophilus alvi gen. nov., sp. nov., a Novel Hydrogenotrophic Methyl-Reducing Methanogenic Archaea of the Order Methanomassiliicoccales Isolated from the Human Gut and Proposal of the Novel Family Methanomethylophilaceae fam. nov
Abstract
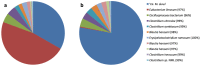
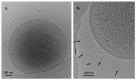
The methanogenic strain Mx-05T was isolated from the human fecal microbiome. A phylogenetic analysis based on the 16S rRNA gene and protein marker genes indicated that the strain is affiliated with the order Methanomassiliicoccales. It shares 86.9% 16S rRNA gene sequence identity with Methanomassiliicoccus luminyensis, the only member of this order previously isolated. The cells of Mx-05T were non-motile cocci, with a diameter range of 0.4-0.7 μm. They grew anaerobically and reduced methanol, monomethylamine, dimethylamine, and trimethylamine into methane, using H2 as an electron donor. H2/CO2, formate, ethanol, and acetate were not used as energy sources. The growth of Mx-05T required an unknown medium factor(s) provided by Eggerthella lenta and present in rumen fluid. Mx-05T grew between 30 °C and 40 °C (optimum 37 °C), over a pH range of 6.9-8.3 (optimum pH 7.5), and between 0.02 and 0.34 mol.L-1 NaCl (optimum 0.12 mol.L-1 NaCl). The genome is 1.67 Mbp with a G+C content of 55.5 mol%. Genome sequence annotation confirmed the absence of the methyl branch of the H4MPT Wood-Ljungdahl pathway, as described for other Methanomassiliicoccales members. Based on an average nucleotide identity analysis, we propose strain Mx-05T as being a novel representative of the order Methanomassiliicoccales, within the novel family Methanomethylophilaceae, for which the name Methanomethylophilus alvi gen. nov, sp. nov. is proposed. The type strain is Mx-05T (JCM 31474T).
Keywords: Methanomassiliicoccales; archaea; human gut microbiota; methanogens.
Conflict of interest statement
P.V. and N.B. are full-time employees of Lesaffre. The other authors report no conflict of interest.
Figures

References
-
- Iino T., Tamaki H., Tamazawa S., Ueno Y., Ohkuma M., Suzuki K., Igarashi Y., Haruta S. Candidatus Methanogranum caenicola: A Novel Methanogen from the Anaerobic Digested Sludge, and Proposal of Methanomassiliicoccaceae fam. nov. and Methanomassiliicoccales ord. nov., for a Methanogenic Lineage of the Class Thermoplasmata. Microbes Environ. 2013;28:244–250. doi: 10.1264/jsme2.ME12189. - DOI - PMC - PubMed
-
- Söllinger A., Schwab C., Weinmaier T., Loy A., Tveit A.T., Schleper C., Urich T. Phylogenetic and Genomic Analysis of Methanomassiliicoccales in Wetlands and Animal Intestinal Tracts Reveals Clade-Specific Habitat Preferences. FEMS Microbiol. Ecol. 2016;92:fiv149. doi: 10.1093/femsec/fiv149. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

