Conformational Stability of the N-Terminal Region of MDM2
- PMID: 38005300
- PMCID: PMC10673428
- DOI: 10.3390/molecules28227578
Conformational Stability of the N-Terminal Region of MDM2
Abstract
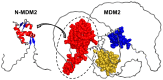
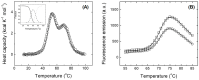
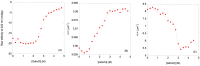
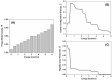
MDM2 is an E3 ubiquitin ligase which is crucial for the degradation and inhibition of the key tumor-suppressor protein p53. In this work, we explored the stability and the conformational features of the N-terminal region of MDM2 (N-MDM2), through which it binds to the p53 protein as well as other protein partners. The isolated domain possessed a native-like conformational stability in a narrow pH range (7.0 to 10.0), as shown by intrinsic and 8-anilinonapthalene-1-sulfonic acid (ANS) fluorescence, far-UV circular dichroism (CD), and size exclusion chromatography (SEC). Guanidinium chloride (GdmCl) denaturation followed by intrinsic and ANS fluorescence, far-UV CD and SEC at physiological pH, and differential scanning calorimetry (DSC) and thermo-fluorescence experiments showed that (i) the conformational stability of isolated N-MDM2 was very low; and (ii) unfolding occurred through the presence of several intermediates. The presence of a hierarchy in the unfolding intermediates was also evidenced through DSC and by simulating the unfolding process with the help of computational techniques based on constraint network analysis (CNA). We propose that the low stability of this protein is related to its inherent flexibility and its ability to interact with several molecular partners through different routes.
Keywords: MDM2; circular dichroism; conformational stability; constraint network analysis; differential scanning calorimetry; fluorescence; intrinsically disordered protein.
Conflict of interest statement
The authors declare no conflict of interest. The authors have no relevant financial or non-financial interests to disclose. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



Similar articles
-
The isolated armadillo-repeat domain of Plakophilin 1 is a monomer in solution with a low conformational stability.J Struct Biol. 2020 Sep 1;211(3):107569. doi: 10.1016/j.jsb.2020.107569. Epub 2020 Jul 7. J Struct Biol. 2020. PMID: 32650131
-
Difference in the mechanisms of the cold and heat induced unfolding of thioredoxin h from Chlamydomonas reinhardtii: spectroscopic and calorimetric studies.Biochemistry. 2000 Sep 12;39(36):11154-62. doi: 10.1021/bi000610b. Biochemistry. 2000. PMID: 10998255
-
Characterization of the equilibrium intermediates in acid denaturation of human stefin B.Eur J Biochem. 1997 Apr 15;245(2):364-72. doi: 10.1111/j.1432-1033.1997.t01-1-00364.x. Eur J Biochem. 1997. PMID: 9151965
-
[Non-native conformational states of immunoglobulins: thermodynamic and functional analysis of rabbit IgG].Biokhimiia. 1996 Feb;61(2):212-35. Biokhimiia. 1996. PMID: 8717493 Review. Russian.
-
Folding/unfolding/refolding of proteins: present methodologies in comparison with capillary zone electrophoresis.Electrophoresis. 2001 Aug;22(12):2359-74. doi: 10.1002/1522-2683(200107)22:12<2359::AID-ELPS2359>3.0.CO;2-8. Electrophoresis. 2001. PMID: 11519938 Review.
References
MeSH terms
Substances
Grants and funding
- PID2021-127296OB-I00 to AVC/Spanish Ministery of Ministry of Science and Innovation MCIN/AEI/10.13039/501100011033/ and "ERDF A way of Making Europe"
- PI18/00394 to OA/Instituto de Salud Carlos III co-funded by European Social Fund "Investing in your future"
- "Protein targets and Bioactive Compounds group" E45-20R to AVC/Diputación General de Aragón
- "Digestive Pathology Group" B25-20R to OA/Diputación General de Aragón
- CIAICO 2021/0135 to JLN/Consellería de Innovación, Universidades, Ciencia y Sociedad Digital (Generalitat Valenciana)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

