Anti-Inflammatory Properties of Eugenol in Lipopolysaccharide-Induced Macrophages and Its Role in Preventing β-Cell Dedifferentiation and Loss Induced by High Glucose-High Lipid Conditions
- PMID: 38005341
- PMCID: PMC10673503
- DOI: 10.3390/molecules28227619
Anti-Inflammatory Properties of Eugenol in Lipopolysaccharide-Induced Macrophages and Its Role in Preventing β-Cell Dedifferentiation and Loss Induced by High Glucose-High Lipid Conditions
Abstract
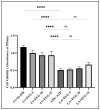
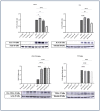
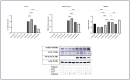
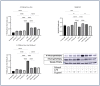
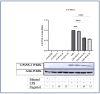
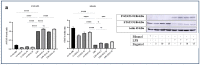
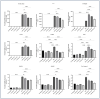
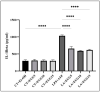
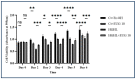
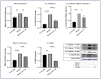
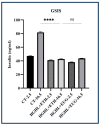
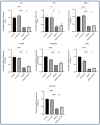
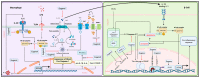
Inflammation is a natural immune response to injury, infection, or tissue damage. It plays a crucial role in maintaining overall health and promoting healing. However, when inflammation becomes chronic and uncontrolled, it can contribute to the development of various inflammatory conditions, including type 2 diabetes. In type 2 diabetes, pancreatic β-cells have to overwork and the continuous impact of a high glucose, high lipid (HG-HL) diet contributes to their loss and dedifferentiation. This study aimed to investigate the anti-inflammatory effects of eugenol and its impact on the loss and dedifferentiation of β-cells. THP-1 macrophages were pretreated with eugenol for one hour and then exposed to lipopolysaccharide (LPS) for three hours to induce inflammation. Additionally, the second phase of NLRP3 inflammasome activation was induced by incubating the LPS-stimulated cells with adenosine triphosphate (ATP) for 30 min. The results showed that eugenol reduced the expression of proinflammatory genes, such as IL-1β, IL-6 and cyclooxygenase-2 (COX-2), potentially by inhibiting the activation of transcription factors NF-κB and TYK2. Eugenol also demonstrated inhibitory effects on the levels of NLRP3 mRNA and protein and Pannexin-1 (PANX-1) activation, eventually impacting the assembly of the NLRP3 inflammasome and the production of mature IL-1β. Additionally, eugenol reduced the elevated levels of adenosine deaminase acting on RNA 1 (ADAR1) transcript, suggesting its role in post-transcriptional mechanisms that regulate inflammatory responses. Furthermore, eugenol effectively decreased the loss of β-cells in response to HG-HL, likely by mitigating apoptosis. It also showed promise in suppressing HG-HL-induced β-cell dedifferentiation by restoring β-cell-specific biomarkers. Further research on eugenol and its mechanisms of action could lead to the development of therapeutic interventions for inflammatory disorders and the preservation of β-cell function in the context of type 2 diabetes.
Keywords: T2DM; cell viability; eugenol; high glucose; high lipid; inflammation; macrophage; β-cell.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- Srinivasan S., Sathish G., Jayanthi M., Muthukumaran J., Muruganathan U., Ramachandran V. Ameliorating effect of eugenol on hyperglycemia by attenuating the key enzymes of glucose metabolism in streptozotocin-induced diabetic rats. Mol. Cell. Biochem. 2014;385:159–168. doi: 10.1007/s11010-013-1824-2. - DOI - PubMed
-
- Holt R.I., Cockram C., Flyvbjerg A., Goldstein B.J. Textbook of Diabetes. John Wiley & Sons; Hoboken, NJ, USA: 2017.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

