Dual-Signal Feature Spaces Map Protein Subcellular Locations Based on Immunohistochemistry Image and Protein Sequence
- PMID: 38005402
- PMCID: PMC10675401
- DOI: 10.3390/s23229014
Dual-Signal Feature Spaces Map Protein Subcellular Locations Based on Immunohistochemistry Image and Protein Sequence
Abstract
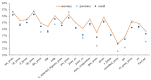
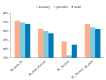
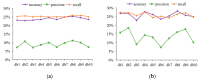
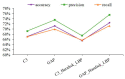
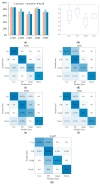
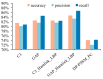
Protein is one of the primary biochemical macromolecular regulators in the compartmental cellular structure, and the subcellular locations of proteins can therefore provide information on the function of subcellular structures and physiological environments. Recently, data-driven systems have been developed to predict the subcellular location of proteins based on protein sequence, immunohistochemistry (IHC) images, or immunofluorescence (IF) images. However, the research on the fusion of multiple protein signals has received little attention. In this study, we developed a dual-signal computational protocol by incorporating IHC images into protein sequences to learn protein subcellular localization. Three major steps can be summarized as follows in this protocol: first, a benchmark database that includes 281 proteins sorted out from 4722 proteins of the Human Protein Atlas (HPA) and Swiss-Prot database, which is involved in the endoplasmic reticulum (ER), Golgi apparatus, cytosol, and nucleoplasm; second, discriminative feature operators were first employed to quantitate protein image-sequence samples that include IHC images and protein sequence; finally, the feature subspace of different protein signals is absorbed to construct multiple sub-classifiers via dimensionality reduction and binary relevance (BR), and multiple confidence derived from multiple sub-classifiers is adopted to decide subcellular location by the centralized voting mechanism at the decision layer. The experimental results indicated that the dual-signal model embedded IHC images and protein sequences outperformed the single-signal models with accuracy, precision, and recall of 75.41%, 80.38%, and 74.38%, respectively. It is enlightening for further research on protein subcellular location prediction under multi-signal fusion of protein.
Keywords: benchmark database; discriminative feature operators; dual signal; protein subcellular location prediction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
Substances
Grants and funding
- 62163017, 61603161/the National Natural Science Foundation of China
- QNJG2020065/the Scholastic Youth Talent Jinggang Program of Jiangxi Province
- 20202BAB202009/the Natural Science Foundation of Jiangxi Province of China
- GJJ160768/the Key Science Foundation of Educational Commission of Jiangxi Province of China
- 2016QNBJRC004/the Scholastic Youth Talent Program of Jiangxi Science and Technology Normal University
LinkOut - more resources
Full Text Sources
Miscellaneous

