Transcriptome Analysis Reveals the Stress Tolerance Mechanisms of Cadmium in Zoysia japonica
- PMID: 38005730
- PMCID: PMC10674853
- DOI: 10.3390/plants12223833
Transcriptome Analysis Reveals the Stress Tolerance Mechanisms of Cadmium in Zoysia japonica
Abstract
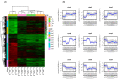
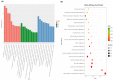
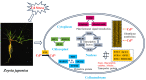
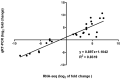
Cadmium (Cd) is a severe heavy metal pollutant globally. Zoysia japonica is an important perennial warm-season turf grass that potentially plays a role in phytoremediation in Cd-polluted soil areas; however, the molecular mechanisms underlying its Cd stress response are unknown. To further investigate the early gene response pattern in Z. japonica under Cd stress, plant leaves were harvested 0, 6, 12, and 24 h after Cd stress (400 μM CdCl2) treatment and used for a time-course RNA-sequencing analysis. Twelve cDNA libraries were constructed and sequenced, and high-quality data were obtained, whose mapped rates were all higher than 94%, and more than 601 million bp of sequence were generated. A total of 5321, 6526, and 4016 differentially expressed genes were identified 6, 12, and 24 h after Cd stress treatment, respectively. A total of 1660 genes were differentially expressed at the three time points, and their gene expression profiles over time were elucidated. Based on the analysis of these genes, the important mechanisms for the Cd stress response in Z. japonica were identified. Specific genes participating in glutathione metabolism, plant hormone signal and transduction, members of protein processing in the endoplasmic reticulum, transporter proteins, transcription factors, and carbohydrate metabolism pathways were further analyzed in detail. These genes may contribute to the improvement of Cd tolerance in Z. japonica. In addition, some candidate genes were highlighted for future studies on Cd stress resistance in Z. japonica and other plants. Our results illustrate the early gene expression response of Z. japonica leaves to Cd and provide some new understanding of the molecular mechanisms of Cd stress in Zosia and Gramineae species.
Keywords: Zoysia japonica; cadmium; differentially expressed genes; molecular mechanism; transcriptome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources

