Exploring Tomato Fruit Viromes through Transcriptome Data Analysis
- PMID: 38005817
- PMCID: PMC10674750
- DOI: 10.3390/v15112139
Exploring Tomato Fruit Viromes through Transcriptome Data Analysis
Abstract
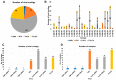
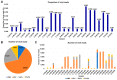
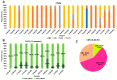
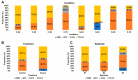
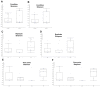
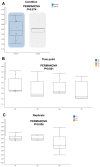
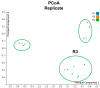
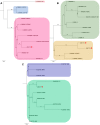
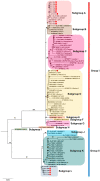
This study delves into the complex landscape of viral infections in tomatoes (Solanum lycopersicum) using available transcriptome data. We conducted a virome analysis, revealing 219 viral contigs linked to four distinct viruses: tomato chlorosis virus (ToCV), southern tomato virus (STV), tomato yellow leaf curl virus (TYLCV), and cucumber mosaic virus (CMV). Among these, ToCV predominated in contig count, followed by STV, TYLCV, and CMV. A notable finding was the prevalence of coinfections, emphasizing the concurrent presence of multiple viruses in tomato plants. Despite generally low viral levels in fruit transcriptomes, STV emerged as the primary virus based on viral read count. We delved deeper into viral abundance and the contributions of RNA segments to replication. While initially focused on studying the impact of sound treatment on tomato fruit transcriptomes, the unexpected viral presence underscores the importance of considering viruses in plant research. Geographical variations in virome communities hint at potential forensic applications. Phylogenetic analysis provided insights into viral origins and genetic diversity, enhancing our understanding of the Korean tomato virome. In conclusion, this study advances our knowledge of the tomato virome, stressing the need for robust pest control in greenhouse-grown tomatoes and offering insights into virus management and crop protection.
Keywords: coinfections; high-throughput sequencing; phylogenetic analysis; tomato fruit virome; viral diversity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Dorais M., Ehret D.L., Papadopoulos A.P. Tomato (Solanum lycopersicum) health components: From the seed to the consumer. Phytochem. Rev. 2008;7:231–250. doi: 10.1007/s11101-007-9085-x. - DOI
-
- Egea I., Estrada Y., Flores F.B., Bolarín M.C. Improving production and fruit quality of tomato under abiotic stress: Genes for the future of tomato breeding for a sustainable agriculture. Environ. Exp. Bot. 2022;204:105086. doi: 10.1016/j.envexpbot.2022.105086. - DOI
-
- Kissoudis C., Chowdhury R., van Heusden S., van de Wiel C., Finkers R., Visser R.G., Bai Y., van der Linden G. Combined biotic and abiotic stress resistance in tomato. Euphytica. 2015;202:317–332. doi: 10.1007/s10681-015-1363-x. - DOI
-
- Marchant W.G., Mugerwa H., Gautam S., Al-Aqeel H., Polston J.E., Rennberger G., Smith H., Turechek B., Adkins S., Brown J.K. Phylogenomic and population genetics analyses of extant tomato yellow leaf curl virus strains on a global scale. Front. Virol. 2023;3:1221156. doi: 10.3389/fviro.2023.1221156. - DOI
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical

