Spike Gene Analysis and Prevalence of Porcine Epidemic Diarrhea Virus from Pigs in South Korea: 2013-2022
- PMID: 38005843
- PMCID: PMC10674705
- DOI: 10.3390/v15112165
Spike Gene Analysis and Prevalence of Porcine Epidemic Diarrhea Virus from Pigs in South Korea: 2013-2022
Abstract
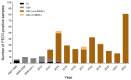
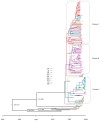
From late 2013-2022, 1131 cases of porcine epidemic diarrhea (PED) were reported to the Korean Animal Health Integrated System (KAHIS). There were four major outbreaks from winter to spring (2013-2014, 2017-2018, 2018-2019, and 2021-2022), with the main outbreaks occurring in Chungnam (CN), Jeonbuk (JB), and Jeju (JJ). Analysis of the complete spike (S) gene of 140/1131 KAHIS PEDV cases nationwide confirmed that 139 belonged to the G2b genotype and 1 to the G2a genotype. Among them, two strains (K17GG1 and K17GB3) were similar to an S INDEL isolated in the United States (strain OH851), and 12 strains had deletions (nucleotides (nt) 3-99) or insertions (12 nt) within the S gene. PEDVs in JJ formed a regionally independent cluster. The substitution rates (substitutions/site/year) were as follows: 1.5952 × 10-3 in CN, 1.8065 × 10-3 in JB, and 1.5113 × 10-3 in JJ. A Bayesian skyline plot showed that the effective population size of PEDs in JJ fell from 2013-2022, whereas in CN and JB it was maintained. Genotyping of 340 Korean PEDV strains, including the 140 PEDVs in this study and 200 Korean reference strains from GenBank, revealed that only the highly pathogenic non-INDEL type (G2b) was dominant from 2020 onwards. Therefore, it is predicted that the incidence of PED will be maintained by the G2b (non-INDEL) genotype.
Keywords: PEDV; S INDEL; diarrhea; evolution; piglet; spike.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Genomic and antigenic characterization of porcine epidemic diarrhoea virus strains isolated from South Korea, 2017.Transbound Emerg Dis. 2018 Aug;65(4):949-956. doi: 10.1111/tbed.12904. Epub 2018 May 16. Transbound Emerg Dis. 2018. PMID: 29770590 Free PMC article.
-
Molecular characteristics and pathogenic assessment of porcine epidemic diarrhoea virus isolates from the 2018 endemic outbreaks on Jeju Island, South Korea.Transbound Emerg Dis. 2019 Sep;66(5):1894-1909. doi: 10.1111/tbed.13219. Epub 2019 May 20. Transbound Emerg Dis. 2019. PMID: 31055885 Free PMC article.
-
Isolation and characterization of Chinese porcine epidemic diarrhea virus with novel mutations and deletions in the S gene.Vet Microbiol. 2018 Jul;221:81-89. doi: 10.1016/j.vetmic.2018.05.021. Epub 2018 Jun 6. Vet Microbiol. 2018. PMID: 29981713 Free PMC article.
-
Complete genome sequences of novel S-deletion variants of porcine epidemic diarrhea virus identified from a recurrent outbreak on Jeju Island, South Korea.Arch Virol. 2019 Oct;164(10):2621-2625. doi: 10.1007/s00705-019-04360-4. Epub 2019 Jul 26. Arch Virol. 2019. PMID: 31350613 Free PMC article.
-
Molecular characterization of US-like and Asian non-S INDEL strains of porcine epidemic diarrhea virus (PEDV) that circulated in Japan during 2013-2016 and PEDVs collected from recurrent outbreaks.BMC Vet Res. 2018 Mar 14;14(1):96. doi: 10.1186/s12917-018-1409-0. BMC Vet Res. 2018. PMID: 29540176 Free PMC article.
Cited by
-
Molecular Characterization of Porcine Epidemic Diarrhea Virus from Field Samples in South Korea.Viruses. 2023 Dec 14;15(12):2428. doi: 10.3390/v15122428. Viruses. 2023. PMID: 38140669 Free PMC article.
-
Targeting pseudoknots with Cas13b inhibits porcine epidemic diarrhoea virus replication.J Gen Virol. 2025 Feb;106(2):002071. doi: 10.1099/jgv.0.002071. J Gen Virol. 2025. PMID: 39903512 Free PMC article.
-
Heat Shock Protein 70 in Cold-Stressed Farm Animals: Implications for Viral Disease Seasonality.Microorganisms. 2025 Jul 27;13(8):1755. doi: 10.3390/microorganisms13081755. Microorganisms. 2025. PMID: 40871259 Free PMC article. Review.
-
In vivo investigation of PEDV transmission via nasal infection: mechanisms of CD4+ T-cell-mediated intestinal infection.J Virol. 2025 Apr 15;99(4):e0176124. doi: 10.1128/jvi.01761-24. Epub 2025 Mar 17. J Virol. 2025. PMID: 40094365 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

