Novel P335-like Phage Resistance Arises from Deletion within Putative Autolysin yccB in Lactococcus lactis
- PMID: 38005870
- PMCID: PMC10675428
- DOI: 10.3390/v15112193
Novel P335-like Phage Resistance Arises from Deletion within Putative Autolysin yccB in Lactococcus lactis
Abstract
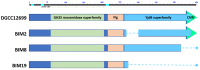
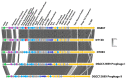
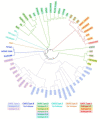
Lactococcus lactis and Lactococcus cremoris are broadly utilized as starter cultures for fermented dairy products and are inherently impacted by bacteriophage (phage) attacks in the industrial environment. Consequently, the generation of bacteriophage-insensitive mutants (BIMs) is a standard approach for addressing phage susceptibility in dairy starter strains. In this study, we characterized spontaneous BIMs of L. lactis DGCC12699 that gained resistance against homologous P335-like phages. Phage resistance was found to result from mutations in the YjdB domain of yccB, a putative autolysin gene. We further observed that alteration of a fused tail-associated lysin-receptor binding protein (Tal-RBP) in the phage restored infectivity on the yccB BIMs. Additional investigation found yccB homologs to be widespread in L. lactis and L. cremoris and that different yccB homologs are highly correlated with cell wall polysaccharide (CWPS) type/subtype. CWPS are known lactococcal phage receptors, and we found that truncation of a glycosyltransferase in the cwps operon also resulted in resistance to these P335-like phages. However, characterization of the CWPS mutant identified notable differences from the yccB mutants, suggesting the two resistance mechanisms are distinct. As phage resistance correlated with yccB mutation has not been previously described in L. lactis, this study offers insight into a novel gene involved in lactococcal phage sensitivity.
Keywords: Lactococcus; autolysin; bacteriophage; resistance; yccB.
Conflict of interest statement
The authors are all employees of International Flavors and Fragrances (IFF), a company that produces and markets cultures for industrial dairy applications.
Figures

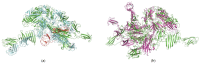
Similar articles
-
PBP2b Mutations Improve the Growth of Phage-Resistant Lactococcus cremoris Lacking Polysaccharide Pellicle.Appl Environ Microbiol. 2023 Jun 28;89(6):e0210322. doi: 10.1128/aem.02103-22. Epub 2023 May 24. Appl Environ Microbiol. 2023. PMID: 37222606 Free PMC article.
-
Differences in lactococcal cell wall polysaccharide structure are major determining factors in bacteriophage sensitivity.mBio. 2014 May 6;5(3):e00880-14. doi: 10.1128/mBio.00880-14. mBio. 2014. PMID: 24803515 Free PMC article.
-
Molecular insights on the recognition of a Lactococcus lactis cell wall pellicle by the phage 1358 receptor binding protein.J Virol. 2014 Jun;88(12):7005-15. doi: 10.1128/JVI.00739-14. Epub 2014 Apr 9. J Virol. 2014. PMID: 24719416 Free PMC article.
-
Interaction between the genomes of Lactococcus lactis and phages of the P335 species.Front Microbiol. 2013 Aug 30;4:257. doi: 10.3389/fmicb.2013.00257. eCollection 2013. Front Microbiol. 2013. PMID: 24009606 Free PMC article.
-
Dairy lactococcal and streptococcal phage-host interactions: an industrial perspective in an evolving phage landscape.FEMS Microbiol Rev. 2020 Nov 24;44(6):909-932. doi: 10.1093/femsre/fuaa048. FEMS Microbiol Rev. 2020. PMID: 33016324 Review.
Cited by
-
Strengthening phage resistance of Streptococcus thermophilus by leveraging complementary defense systems.Nat Commun. 2025 Aug 4;16(1):7142. doi: 10.1038/s41467-025-62408-3. Nat Commun. 2025. PMID: 40759662 Free PMC article.
References
-
- Coffey, Coakley, McGarry, Fitzgerald, Ross Increasing phage resistance of cheese starters: A case study using Lactococcus lactis DPC4268. Lett. Appl. Microbiol. 2002;26:51–55. doi: 10.1046/j.1472-765X.1998.00268.x. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

