Human Complement Inhibits Myophages against Pseudomonas aeruginosa
- PMID: 38005888
- PMCID: PMC10674969
- DOI: 10.3390/v15112211
Human Complement Inhibits Myophages against Pseudomonas aeruginosa
Abstract
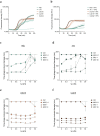
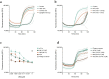
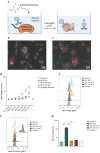
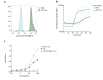
Therapeutic bacteriophages (phages) are primarily chosen based on their in vitro bacteriolytic activity. Although anti-phage antibodies are known to inhibit phage infection, the influence of other immune system components is less well known. An important anti-bacterial and anti-viral innate immune system that may interact with phages is the complement system, a cascade of proteases that recognizes and targets invading microorganisms. In this research, we aimed to study the effects of serum components such as complement on the infectivity of different phages targeting Pseudomonas aeruginosa. We used a fluorescence-based assay to monitor the killing of P. aeruginosa by phages of different morphotypes in the presence of human serum. Our results reveal that several myophages are inhibited by serum in a concentration-dependent way, while the activity of four podophages and one siphophage tested in this study is not affected by serum. By using specific nanobodies blocking different components of the complement cascade, we showed that activation of the classical complement pathway is a driver of phage inhibition. To determine the mechanism of inhibition, we produced bioorthogonally labeled fluorescent phages to study their binding by means of microscopy and flow cytometry. We show that phage adsorption is hampered in the presence of active complement. Our results indicate that interactions with complement may affect the in vivo activity of therapeutically administered phages. A better understanding of this phenomenon is essential to optimize the design and application of therapeutic phage cocktails.
Keywords: Pseudomonas aeruginosa; complement system; phage therapy.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Similar articles
-
Relevance of the bacteriophage adherence to mucus model for Pseudomonas aeruginosa phages.Microbiol Spectr. 2024 Aug 6;12(8):e0352023. doi: 10.1128/spectrum.03520-23. Epub 2024 Jun 24. Microbiol Spectr. 2024. PMID: 38912817 Free PMC article.
-
Systematic bacteriophage selection for the lysis of multiple Pseudomonas aeruginosa strains.Front Cell Infect Microbiol. 2025 May 23;15:1597009. doi: 10.3389/fcimb.2025.1597009. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40487314 Free PMC article.
-
Two Novel Bacteriophages Improve Survival in Galleria mellonella Infection and Mouse Acute Pneumonia Models Infected with Extensively Drug-Resistant Pseudomonas aeruginosa.Appl Environ Microbiol. 2019 Apr 18;85(9):e02900-18. doi: 10.1128/AEM.02900-18. Print 2019 May 1. Appl Environ Microbiol. 2019. PMID: 30824445 Free PMC article.
-
Bacteriophages of Pseudomonas aeruginosa: long-term prospects for use in phage therapy.Adv Virus Res. 2014;88:227-78. doi: 10.1016/B978-0-12-800098-4.00005-2. Adv Virus Res. 2014. PMID: 24373314 Review.
-
Phage Therapy: a Step Forward in the Treatment of Pseudomonas aeruginosa Infections.J Virol. 2015 Aug;89(15):7449-56. doi: 10.1128/JVI.00385-15. Epub 2015 May 13. J Virol. 2015. PMID: 25972556 Free PMC article. Review.
Cited by
-
A Novel Bacteriophage Infecting Multi-Drug- and Extended-Drug-Resistant Pseudomonas aeruginosa Strains.Antibiotics (Basel). 2024 Jun 3;13(6):523. doi: 10.3390/antibiotics13060523. Antibiotics (Basel). 2024. PMID: 38927189 Free PMC article.
-
The contribution of neutrophils to bacteriophage clearance and pharmacokinetics in vivo.JCI Insight. 2024 Oct 22;9(20):e181309. doi: 10.1172/jci.insight.181309. JCI Insight. 2024. PMID: 39435664 Free PMC article.
-
The complement system: A key player in the host response to infections.Eur J Immunol. 2024 Nov;54(11):e2350814. doi: 10.1002/eji.202350814. Epub 2024 Aug 27. Eur J Immunol. 2024. PMID: 39188171 Review.
References
-
- Ferry T., Kolenda C., Laurent F., Leboucher G., Merabischvilli M., Djebara S., Gustave C.-A., Perpoint T., Barrey C., Pirnay J.-P., et al. Personalized Bacteriophage Therapy to Treat Pandrug-Resistant Spinal Pseudomonas aeruginosa Infection. Nat. Commun. 2022;13:4239. doi: 10.1038/s41467-022-31837-9. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

