In Vivo and In Vitro Characterization of the Recently Emergent PRRSV 1-4-4 L1C Variant (L1C.5) in Comparison with Other PRRSV-2 Lineage 1 Isolates
- PMID: 38005910
- PMCID: PMC10674456
- DOI: 10.3390/v15112233
In Vivo and In Vitro Characterization of the Recently Emergent PRRSV 1-4-4 L1C Variant (L1C.5) in Comparison with Other PRRSV-2 Lineage 1 Isolates
Abstract
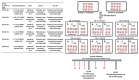
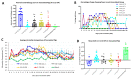
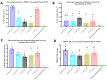
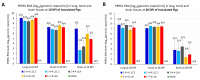
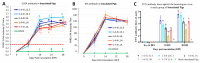
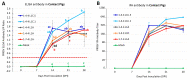
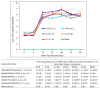
The recently emerged PRRSV 1-4-4 L1C variant (L1C.5) was in vivo and in vitro characterized in this study in comparison with three other contemporary 1-4-4 isolates (L1C.1, L1A, and L1H) and one 1-7-4 L1A isolate. Seventy-two 3-week-old PRRSV-naive pigs were divided into six groups with twelve pigs/group. Forty-eight pigs (eight/group) were for inoculation, and 24 pigs (four/group) served as contact pigs. Pigs in pen A of each room were inoculated with the corresponding virus or negative media. At two days post inoculation (DPI), contact pigs were added to pen B adjacent to pen A in each room. Pigs were necropsied at 10 and 28 DPI. Compared to other virus-inoculated groups, the L1C.5-inoculated pigs exhibited more severe anorexia and lethargy, higher mortality, a higher fraction of pigs with fever (>40 °C), higher average temperature at several DPIs, and higher viremia levels at 2 DPI. A higher percentage of the contact pigs in the L1C.5 group became viremic at two days post contact, implying the higher transmissibility of this virus strain. It was also found that some PRRSV isolates caused brain infection in inoculation pigs and/or contact pigs. The complete genome sequences and growth characteristics in ZMAC cells of five PRRSV-2 isolates were further compared. Collectively, this study confirms that the PRRSV 1-4-4 L1C variant (L1C.5) is highly virulent with potential higher transmissibility, but the genetic determinants of virulence remain to be elucidated.
Keywords: 1-4-4 L1C variant; L1C.1; L1C.5; PRRSV; brain infection; experimental study; porcine reproductive and respiratory syndrome virus; transmissibility; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Fang Y., Treffers E.E., Li Y., Tas A., Sun Z., van der Meer Y., de Ru A.H., van Veelen P.A., Atkins J.F., Snijder E.J., et al. Efficient -2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc. Natl. Acad. Sci. USA. 2012;109:E2920–E2928. doi: 10.1073/pnas.1211145109. - DOI - PMC - PubMed
-
- Zimmerman J., Dee S., Holtkamp D.J., Murtaugh M., Stadejek T., Stevenson G., Torremorell M., Yang H., Zhang J. Diseases of Swine. 11th ed. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2019. Porcine reproductive and respiratory syndrome viruses (porcine arteriviruses) pp. 685–708.
-
- Yim-im W., Anderson T.K., Paploski I., VanderWaal K., Gauger P., Kreuger K., Shi M., Main R., Zhang J. Refining PRRSV-2 genetic classification based on global ORF5 sequences and investigation of their geographic distributions and temporal changes. Microbiol. Spectr. 2023:e0291623. doi: 10.1128/spectrum.02916-23. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

