In Vivo Validation of Novel Synthetic tbp1 Peptide-Based Vaccine Candidates against Haemophilus influenzae Strains in BALB/c Mice
- PMID: 38005983
- PMCID: PMC10675187
- DOI: 10.3390/vaccines11111651
In Vivo Validation of Novel Synthetic tbp1 Peptide-Based Vaccine Candidates against Haemophilus influenzae Strains in BALB/c Mice
Abstract
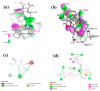
Haemophilus influenzae is a Gram-negative bacterium characterized as a small, nonmotile, facultative anaerobic coccobacillus. It is a common cause of a variety of invasive and non-invasive infections. Among six serotypes (a-f), H. influenzae type b (Hib) is the most familiar and predominant mostly in children and immunocompromised individuals. Following Hib vaccination, infections due to other serotypes have increased in number, and currently, there is no suitable effective vaccine to induce cross-strain protective antibody responses. The current study was aimed to validate the capability of two 20-mer highly conserved synthetic tbp1 (transferrin-binding protein 1) peptide-based vaccine candidates (tbp1-E1 and tbp1-E2) predicted using in silico approaches to induce immune responses against H. influenzae strains. Cytokine induction ability, immune simulations, and molecular dynamics (MD) simulations were performed to confirm the candidacy of epitopic docked complexes. Synthetic peptide vaccine formulations in combination with two different adjuvants, BGs (Bacterial Ghosts) and CFA/IFA (complete/incomplete Freund's adjuvant), were used in BALB/c mouse groups in three booster shots at two-week intervals. An indirect ELISA was performed to determine endpoint antibody titers using the Student's t-distribution method. The results revealed that the synergistic use of both peptides in combination with BG adjuvants produced better results. Significant differences in absorbance values were observed in comparison to the rest of the peptide-adjuvant combinations. The findings of this study indicate that these tbp1 peptide-based vaccine candidates may present a preliminary set of peptides for the development of an effective cross-strain vaccine against H. influenzae in the future due to their highly conserved nature.
Keywords: BALB/c mice; H. influenzae; IgG antibodies; adjuvants; indirect ELISA; peptide antigens; tbp1 peptides.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



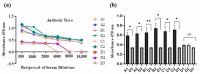
Similar articles
-
Conservation and antigenic cross-reactivity of the transferrin-binding proteins of Haemophilus influenzae, Actinobacillus pleuropneumoniae and Neisseria meningitidis.Microbiology (Reading). 1996 Dec;142 ( Pt 12):3505-13. doi: 10.1099/13500872-142-12-3505. Microbiology (Reading). 1996. PMID: 9004513
-
[Mouse macrophage-derived chemokine as an adjuvant enhances the protective effect of P6 protein vaccine of nontypeable Haemophilus influenzae].Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2014 Sep;30(9):913-6. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2014. PMID: 25200152 Chinese.
-
Cloning and expression of the Haemophilus influenzae transferrin receptor genes.Mol Microbiol. 1996 Feb;19(3):575-86. doi: 10.1046/j.1365-2958.1996.406943.x. Mol Microbiol. 1996. PMID: 8830248
-
Invasive Haemophilus influenzae Infections after 3 Decades of Hib Protein Conjugate Vaccine Use.Clin Microbiol Rev. 2021 Jun 16;34(3):e0002821. doi: 10.1128/CMR.00028-21. Epub 2021 Jun 2. Clin Microbiol Rev. 2021. PMID: 34076491 Free PMC article. Review.
-
Haemophilus influenzae type b disease in the era of conjugate vaccines: critical factors for successful eradication.Expert Rev Vaccines. 2020 Oct;19(10):903-917. doi: 10.1080/14760584.2020.1825948. Epub 2020 Oct 10. Expert Rev Vaccines. 2020. PMID: 32962476 Review.
Cited by
-
Construction and Mechanism Exploration of Highly Efficient System for Bacterial Ghosts Preparation Based on Engineered Phage ID52 Lysis Protein E.Vaccines (Basel). 2024 Apr 28;12(5):472. doi: 10.3390/vaccines12050472. Vaccines (Basel). 2024. PMID: 38793723 Free PMC article.
References
-
- Soeters H.M., Blain A., Pondo T., Monica B.D., Farley M., Harrison L.H., Lynfield R., Miller L., Petit S., Reingold A., et al. Current Epidemiology and Trends in Invasive Haemophilus influenzae Disease—United States, 2009–2015. Clin. Infect. Dis. 2018;67:881–889. doi: 10.1093/cid/ciy187. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

