Comprehensive analysis of scRNA-Seq and bulk RNA-Seq data reveals dynamic changes in tumor-associated neutrophils in the tumor microenvironment of hepatocellular carcinoma and leads to the establishment of a neutrophil-related prognostic model
- PMID: 38006433
- PMCID: PMC10992459
- DOI: 10.1007/s00262-023-03567-4
Comprehensive analysis of scRNA-Seq and bulk RNA-Seq data reveals dynamic changes in tumor-associated neutrophils in the tumor microenvironment of hepatocellular carcinoma and leads to the establishment of a neutrophil-related prognostic model
Abstract
Background: Analysis of hepatocellular carcinoma (HCC) single-cell sequencing data was conducted to explore the role of tumor-associated neutrophils in the tumor microenvironment.
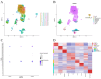
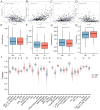
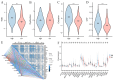
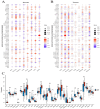
Methods: Analysis of single-cell sequencing data from 12 HCC tumor cores and five HCC paracancerous tissues identified cellular subpopulations and cellular marker genes. The Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus (GEO) databases were used to establish and validate prognostic models. xCELL, TIMER, QUANTISEQ, CIBERSORT, and CIBERSORT-abs analyses were performed to explore immune cell infiltration. Finally, the pattern of tumor-associated neutrophil roles in tumor microenvironmental components was explored.
Results: A total of 271 marker genes for tumor-associated neutrophils were identified based on single-cell sequencing data. Prognostic models incorporating eight genes were established based on TCGA data. Immune cell infiltration differed between the high- and low-risk groups. The low-risk group benefited more from immunotherapy. Single-cell analysis indicated that tumor-associated neutrophils were able to influence macrophage, NK cell, and T-cell functions through the IL16, IFN-II, and SPP1 signaling pathways.
Conclusion: Tumor-associated neutrophils regulate immune functions by influencing macrophages and NK cells. Models incorporating tumor-associated neutrophil-related genes can be used to predict patient prognosis and immunotherapy responses.
Keywords: CellChat; HCC; Immunotherapy; Single-cell; Tumor-associated neutrophils.
© 2023. The Author(s), under exclusive licence to Springer-Verlag GmbH Germany, part of Springer Nature.
Conflict of interest statement
None.
Figures



Similar articles
-
Single-cell and bulk transcriptomic datasets enable the development of prognostic models based on dynamic changes in the tumor immune microenvironment in patients with hepatocellular carcinoma and portal vein tumor thrombus.Front Immunol. 2024 Oct 28;15:1414121. doi: 10.3389/fimmu.2024.1414121. eCollection 2024. Front Immunol. 2024. PMID: 39530087 Free PMC article.
-
Comprehensive scRNA-seq Analysis and Identification of CD8_+T Cell Related Gene Markers for Predicting Prognosis and Drug Resistance of Hepatocellular Carcinoma.Curr Med Chem. 2024;31(17):2414-2430. doi: 10.2174/0109298673274578231030065454. Curr Med Chem. 2024. PMID: 37936457
-
Single-cell sequencing combined with bulk RNA seq reveals the roles of natural killer cell in prognosis and immunotherapy of hepatocellular carcinoma.Sci Rep. 2025 May 1;15(1):15314. doi: 10.1038/s41598-025-99638-w. Sci Rep. 2025. PMID: 40312525 Free PMC article.
-
Harnessing neutrophil plasticity for HCC immunotherapy.Essays Biochem. 2023 Sep 28;67(6):941-955. doi: 10.1042/EBC20220245. Essays Biochem. 2023. PMID: 37534829 Free PMC article. Review.
-
Single-cell RNA sequencing reveals the landscape of the cellular ecosystem of primary hepatocellular carcinoma.Cancer Cell Int. 2024 Nov 14;24(1):379. doi: 10.1186/s12935-024-03574-0. Cancer Cell Int. 2024. PMID: 39543644 Free PMC article. Review.
Cited by
-
Integrative analysis of single-cell and bulk RNA sequencing reveals the oncogenic role of ANXA5 in gastric cancer and its association with drug resistance.Front Immunol. 2025 Mar 7;16:1562395. doi: 10.3389/fimmu.2025.1562395. eCollection 2025. Front Immunol. 2025. PMID: 40124374 Free PMC article.
-
Genomic and Immune Profiling of Esophageal Squamous Cell Carcinoma Undergoing Neoadjuvant Therapy Versus Upfront Surgery Identifies Novel Immunogenic Cell Death-Based Signatures for Predicting Clinical Outcomes.MedComm (2020). 2025 Apr 2;6(4):e70171. doi: 10.1002/mco2.70171. eCollection 2025 Apr. MedComm (2020). 2025. PMID: 40182138 Free PMC article.
-
Integrating single-cell transcriptomics and machine learning to predict breast cancer prognosis: A study based on natural killer cell-related genes.J Cell Mol Med. 2024 Aug;28(15):e18549. doi: 10.1111/jcmm.18549. J Cell Mol Med. 2024. PMID: 39098994 Free PMC article.
-
Identification and assessment of differentially expressed necroptosis long non-coding RNAs associated with periodontitis in human.BMC Oral Health. 2023 Sep 4;23(1):632. doi: 10.1186/s12903-023-03308-0. BMC Oral Health. 2023. Retraction in: BMC Oral Health. 2024 Nov 8;24(1):1359. doi: 10.1186/s12903-024-05157-x. PMID: 37667236 Free PMC article. Retracted.
-
Global trends in tumor-associated neutrophil research: a bibliometric and visual analysis.Front Immunol. 2025 Mar 14;16:1478092. doi: 10.3389/fimmu.2025.1478092. eCollection 2025. Front Immunol. 2025. PMID: 40160822 Free PMC article.
References
-
- Liu Z, Jiang Y, Yuan H, Fang Q, Cai N, Suo C, et al. The trends in incidence of primary liver cancer caused by specific etiologies: results from the global burden of disease study 2016 and implications for liver cancer prevention. J Hepatol. 2019;70(4):674–683. doi: 10.1016/j.jhep.2018.12.001. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

