Single-cell RNA-sequencing atlas reveals an FABP1-dependent immunosuppressive environment in hepatocellular carcinoma
- PMID: 38007237
- PMCID: PMC10679975
- DOI: 10.1136/jitc-2023-007030
Single-cell RNA-sequencing atlas reveals an FABP1-dependent immunosuppressive environment in hepatocellular carcinoma
Abstract
Background: Single-cell RNA sequencing, also known as scRNA-seq, is a method profiling cell populations on an individual cell basis. It is particularly useful for more deeply understanding cell behavior in a complicated tumor microenvironment. Although several previous studies have examined scRNA-seq for hepatocellular carcinoma (HCC) tissues, no one has tested and analyzed HCC with different stages.
Methods: In this investigation, immune cells isolated from surrounding normal tissues and cancer tissues from 3 II-stage and 4 III-stage HCC cases were subjected to deep scRNA-seq. The analysis included 15 samples. We distinguished developmentally relevant trajectories, unique immune cell subtypes, and enriched pathways regarding differential genes. Western blot and co-immunoprecipitation were performed to demonstrate the interaction between fatty acid binding protein 1 (FABP1) and peroxisome proliferator-activated receptor gamma(PPARG). In vivo experiments were performed in a C57BL/6 mouse model of HCC established via subcutaneous injection.
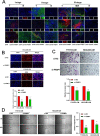
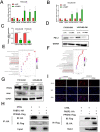
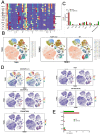
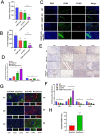
Results: FABP1 was discovered to be overexpressed in tumor-associated macrophages (TAMs) with III-stage HCC tissues compared with II-stage HCC tissues. This finding was fully supported by immunofluorescence detection in significant amounts of HCC human samples. FABP1 deficiency in TAMs inhibited HCC progression in vitro. Mechanistically, FABP1 interacted with PPARG/CD36 in TAMs to increase fatty acid oxidation in HCC. When compared with C57BL/6 mice of the wild type, tumors in FABP1-/- mice consistently showed attenuation. The FABP1-/- group's relative proportion of regulatory T cells and natural killer cells showed a downward trend, while dendritic cells, M1 macrophages, and B cells showed an upward trend, according to the results of mass cytometry. In further clinical translation, we found that orlistat significantly inhibited FABP1 activity, while the combination of anti-programmed cell death 1(PD-1) could synergistically treat HCC progression. Liposomes loaded with orlistat and connected with IR780 probe could further enhance the therapeutic effect of orlistat and visualize drug metabolism in vivo.
Conclusions: ScRNA-seq atlas revealed an FABP1-dependent immunosuppressive environment in HCC. Orlistat significantly inhibited FABP1 activity, while the combination of anti-PD-1 could synergistically treat HCC progression. This study identified new treatment targets and strategies for HCC progression, contributing to patients with advanced HCC from new perspectives.
Keywords: antibodies, neoplasm; antineoplastic protocols; liver neoplasms.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: No, there are no competing interests.
Figures





Similar articles
-
FABP5+ lipid-loaded macrophages process tumour-derived unsaturated fatty acid signal to suppress T-cell antitumour immunity.J Hepatol. 2025 Apr;82(4):676-689. doi: 10.1016/j.jhep.2024.09.029. Epub 2024 Sep 30. J Hepatol. 2025. PMID: 39357545
-
Inhibition of APOC1 promotes the transformation of M2 into M1 macrophages via the ferroptosis pathway and enhances anti-PD1 immunotherapy in hepatocellular carcinoma based on single-cell RNA sequencing.Redox Biol. 2022 Oct;56:102463. doi: 10.1016/j.redox.2022.102463. Epub 2022 Sep 2. Redox Biol. 2022. PMID: 36108528 Free PMC article.
-
TREM2+ macrophages suppress CD8+ T-cell infiltration after transarterial chemoembolisation in hepatocellular carcinoma.J Hepatol. 2023 Jul;79(1):126-140. doi: 10.1016/j.jhep.2023.02.032. Epub 2023 Mar 6. J Hepatol. 2023. PMID: 36889359
-
Crosstalk between tumor-associated macrophages and neighboring cells in hepatocellular carcinoma.Clin Mol Hepatol. 2022 Jul;28(3):333-350. doi: 10.3350/cmh.2021.0308. Epub 2021 Oct 19. Clin Mol Hepatol. 2022. PMID: 34665953 Free PMC article. Review.
-
Understanding initiation and progression of hepatocellular carcinoma through single cell sequencing.Biochim Biophys Acta Rev Cancer. 2022 May;1877(3):188720. doi: 10.1016/j.bbcan.2022.188720. Epub 2022 Mar 15. Biochim Biophys Acta Rev Cancer. 2022. PMID: 35304295 Review.
Cited by
-
Applications of single-cell multi-omics in liver cancer.JHEP Rep. 2024 Apr 15;6(7):101094. doi: 10.1016/j.jhepr.2024.101094. eCollection 2024 Jul. JHEP Rep. 2024. PMID: 39022385 Free PMC article. Review.
-
Single-cell and spatial atlas of glioblastoma heterogeneity: characterizing the PCLAF+ subtype and YEATS4's oncogenic role.Front Immunol. 2025 Jul 25;16:1614549. doi: 10.3389/fimmu.2025.1614549. eCollection 2025. Front Immunol. 2025. PMID: 40787449 Free PMC article.
-
Drug functional remapping: a new promise for tumor immunotherapy.Front Oncol. 2025 Mar 14;15:1519355. doi: 10.3389/fonc.2025.1519355. eCollection 2025. Front Oncol. 2025. PMID: 40161377 Free PMC article. Review.
-
Gastric cancer adapts high lipid microenvironment via suppressing PPARG-FABP1 axis after arriving in the lymph node.Redox Biol. 2025 Jul 17;85:103759. doi: 10.1016/j.redox.2025.103759. Online ahead of print. Redox Biol. 2025. PMID: 40694956 Free PMC article.
-
Lipid Metabolism Reprogramming in Tumor-Associated Macrophages Modulates Their Function in Primary Liver Cancers.Cancers (Basel). 2025 May 31;17(11):1858. doi: 10.3390/cancers17111858. Cancers (Basel). 2025. PMID: 40507339 Free PMC article. Review.
References
-
- European Association for the Study of the Liver. Electronic address eee, European Association for the Study of the L . EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol 2018;69:182–236. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
