Framework humanization optimizes potency of anti-CD72 nanobody CAR-T cells for B-cell malignancies
- PMID: 38007238
- PMCID: PMC10680002
- DOI: 10.1136/jitc-2023-006985
Framework humanization optimizes potency of anti-CD72 nanobody CAR-T cells for B-cell malignancies
Abstract
Background: Approximately 50% of patients who receive anti-CD19 CAR-T cells relapse, and new immunotherapeutic targets are urgently needed. We recently described CD72 as a promising target in B-cell malignancies and developed nanobody-based CAR-T cells (nanoCARs) against it. This cellular therapy design is understudied compared with scFv-based CAR-T cells, but has recently become of significant interest given the first regulatory approval of a nanoCAR in multiple myeloma.
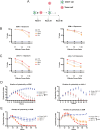
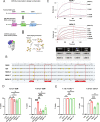
Methods: We humanized our previous nanobody framework regions, derived from llama, to generate a series of humanized anti-CD72 nanobodies. These nanobody binders were inserted into second-generation CD72 CAR-T cells and were evaluated against preclinical models of B cell acute lymphoblastic leukemia and B cell non-Hodgkin's lymphoma in vitro and in vivo. Humanized CD72 nanoCARs were compared with parental ("NbD4") CD72 nanoCARs and the clinically approved CD19-directed CAR-T construct tisangenlecleucel. RNA-sequencing, flow cytometry, and cytokine secretion profiling were used to determine differences between the different CAR constructs. We then used affinity maturation on the parental NbD4 construct to generate high affinity binders against CD72 to test if higher affinity to CD72 improved antitumor potency.
Results: Toward clinical translation, here we humanize our previous nanobody framework regions, derived from llama, and surprisingly discover a clone ("H24") with enhanced potency against B-cell tumors, including patient-derived samples after CD19 CAR-T relapse. Potentially underpinning improved potency, H24 has moderately higher binding affinity to CD72 compared with a fully llama framework. However, further affinity maturation (KD<1 nM) did not lead to improvement in cytotoxicity. After treatment with H24 nanoCARs, in vivo relapse was accompanied by CD72 antigen downregulation which was partially reversible. The H24 nanobody clone was found to have no off-target binding and is therefore designated as a true clinical candidate.
Conclusion: This work supports translation of H24 CD72 nanoCARs for refractory B-cell malignancies, reveals potential mechanisms of resistance, and unexpectedly demonstrates that nanoCAR potency can be improved by framework alterations alone. These findings may have implications for future engineering of nanobody-based cellular therapies.
Keywords: Cell Engineering; Hematologic Neoplasms; Immunotherapy; Receptors, Chimeric Antigen; Translational Medical Research.
© Author(s) (or their employer(s)) 2023. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: APW and MAN have filed intellectual property claims relevant to the nanobody sequences described here. MAN is an employee and equity shareholder of Cartography Biosciences. APW has received research funding from Genentech/Roche. The other authors declare no relevant conflicts of interest.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
