Paneth cell-derived iNOS is required to maintain homeostasis in the intestinal stem cell niche
- PMID: 38007452
- PMCID: PMC10675917
- DOI: 10.1186/s12967-023-04744-w
Paneth cell-derived iNOS is required to maintain homeostasis in the intestinal stem cell niche
Abstract
Background: Mammalian intestinal epithelium constantly undergoes rapid self-renewal and regeneration sustained by intestinal stem cells (ISCs) within crypts. Inducible nitric oxide synthase (iNOS) is an important regulator in tissue homeostasis and inflammation. However, the functions of iNOS on ISCs have not been clarified. Here, we aimed to investigate the expression pattern of inducible nitric oxide synthase (iNOS) within crypts and explore its function in the homeostatic maintenance of the ISC niche.
Methods: Expression of iNOS was determined by tissue staining and qPCR. iNOS-/- and Lgr5 transgenic mice were used to explore the influence of iNOS ablation on ISC proliferation and differentiation. Enteroids were cultured to study the effect of iNOS on ISCs in vitro. Ileum samples from wild-type and iNOS-/- mice were collected for RNA-Seq to explore the molecular mechanisms by which iNOS regulates ISCs.
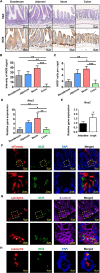
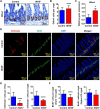
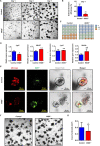
Results: iNOS was physiologically expressed in Paneth cells. Knockout of iNOS led to apparent morphological changes in the intestine, including a decrease in the small intestine length and in the heights of both villi and crypts. Knockout of iNOS decreased the number of Ki67+ or BrdU+ proliferative cells in crypts. Loss of iNOS increased the number of Olfm4+ ISCs but inhibited the differentiation and migration of Lgr5+ ISCs in vivo. iNOS depletion also inhibited enteroid formation and the budding efficiency of crypts in vitro. Moreover, iNOS deficiency altered gluconeogenesis and the adaptive immune response in the ileum transcriptome.
Conclusion: Paneth cell-derived iNOS is required to maintain a healthy ISC niche, and Knockout of iNOS hinders ISC function in mice. Therefore, iNOS represents a potential target for the development of new drugs and other therapeutic interventions for intestinal disorders.
Keywords: Differentiation; Intestinal stem cell; Paneth cell; Proliferation; iNOS.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
Simultaneous real-time analysis of Paneth cell and intestinal stem cell response to interferon-γ by a novel stem cell niche tracking method.Biochem Biophys Res Commun. 2021 Mar 19;545:14-19. doi: 10.1016/j.bbrc.2021.01.050. Epub 2021 Jan 30. Biochem Biophys Res Commun. 2021. PMID: 33529805
-
The phosphatase PRL-3 affects intestinal homeostasis by altering the crypt cell composition.J Mol Med (Berl). 2021 Oct;99(10):1413-1426. doi: 10.1007/s00109-021-02097-9. Epub 2021 Jun 15. J Mol Med (Berl). 2021. PMID: 34129057 Free PMC article.
-
Chronic GPER activation prompted the proliferation of ileal stem cell in ovariectomized mice depending on Paneth cell-derived Wnt3.Clin Sci (Lond). 2023 Jan 13;137(1):109-127. doi: 10.1042/CS20220392. Clin Sci (Lond). 2023. PMID: 36503938
-
Paneth Cells: Dispensable yet Irreplaceable for the Intestinal Stem Cell Niche.Cell Mol Gastroenterol Hepatol. 2025;19(4):101443. doi: 10.1016/j.jcmgh.2024.101443. Epub 2024 Dec 19. Cell Mol Gastroenterol Hepatol. 2025. PMID: 39708920 Free PMC article. Review.
-
The Intestinal Stem Cell Niche: Homeostasis and Adaptations.Trends Cell Biol. 2018 Dec;28(12):1062-1078. doi: 10.1016/j.tcb.2018.08.001. Epub 2018 Sep 5. Trends Cell Biol. 2018. PMID: 30195922 Free PMC article. Review.
Cited by
-
Gut aging: A wane from the normal to repercussion and gerotherapeutic strategies.Heliyon. 2024 Sep 12;10(19):e37883. doi: 10.1016/j.heliyon.2024.e37883. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39381110 Free PMC article. Review.
-
Cooperation of Wnt/β-catenin and Dll1-mediated Notch pathway in Lgr5-positive intestinal stem cells regulates the mucosal injury and repair in DSS-induced colitis mice model.Gastroenterol Rep (Oxf). 2024 Oct 23;12:goae090. doi: 10.1093/gastro/goae090. eCollection 2024. Gastroenterol Rep (Oxf). 2024. PMID: 39444950 Free PMC article.
-
Novel breast reconstruction technique using ex vivo mononuclear (RE-01) cells and adipose-derived mesenchymal stem cells.Regen Ther. 2025 Mar 29;29:271-281. doi: 10.1016/j.reth.2025.03.018. eCollection 2025 Jun. Regen Ther. 2025. PMID: 40230355 Free PMC article.
-
Recognizing the biological barriers and pathophysiological characteristics of the gastrointestinal tract for the design and application of nanotherapeutics.Drug Deliv. 2024 Dec;31(1):2415580. doi: 10.1080/10717544.2024.2415580. Epub 2024 Oct 15. Drug Deliv. 2024. PMID: 39404464 Free PMC article. Review.
-
Circulating lung cancer exosomes damage the niche of intestinal stem cells.Transl Lung Cancer Res. 2025 Mar 31;14(3):718-735. doi: 10.21037/tlcr-24-758. Epub 2025 Mar 10. Transl Lung Cancer Res. 2025. PMID: 40248740 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

