Genetic diversity and historical demography of underutilised goat breeds in North-Western Europe
- PMID: 38007600
- PMCID: PMC10676416
- DOI: 10.1038/s41598-023-48005-8
Genetic diversity and historical demography of underutilised goat breeds in North-Western Europe
Abstract
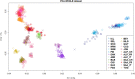
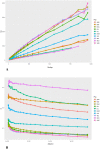
In the last decade, several studies aimed at dissecting the genetic architecture of local small ruminant breeds to discover which variations are involved in the process of adaptation to environmental conditions, a topic that has acquired priority due to climate change. Considering that traditional breeds are a reservoir of such important genetic variation, improving the current knowledge about their genetic diversity and origin is the first step forward in designing sound conservation guidelines. The genetic composition of North-Western European archetypical goat breeds is still poorly exploited. In this study we aimed to fill this gap investigating goat breeds across Ireland and Scandinavia, including also some other potential continental sources of introgression. The PCA and Admixture analyses suggest a well-defined cluster that includes Norwegian and Swedish breeds, while the crossbred Danish landrace is far apart, and there appears to be a close relationship between the Irish and Saanen goats. In addition, both graph representation of historical relationships among populations and f4-ratio statistics suggest a certain degree of gene flow between the Norse and Atlantic landraces. Furthermore, we identify signs of ancient admixture events of Scandinavian origin in the Irish and in the Icelandic goats. The time when these migrations, and consequently the introgression, of Scandinavian-like alleles occurred, can be traced back to the Viking colonisation of these two isles during the Viking Age (793-1066 CE). The demographic analysis indicates a complicated history of these traditional breeds with signatures of bottleneck, inbreeding and crossbreeding with the improved breeds. Despite these recent demographic changes and the historical genetic background shaped by centuries of human-mediated gene flow, most of them maintained their genetic identity, becoming an irreplaceable genetic resource as well as a cultural heritage.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Macheridis S. Animal husbandry in Iron Age Scania, with a catalogue. Acta Archaeol. Lundensia Ser. Altera. 2022;73:263.
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

