Identifying phenotype-associated subpopulations through LP_SGL
- PMID: 38008419
- PMCID: PMC10753413
- DOI: 10.1093/bib/bbad424
Identifying phenotype-associated subpopulations through LP_SGL
Abstract
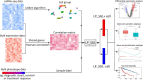
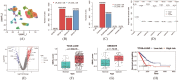
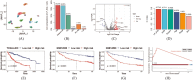
Single-cell RNA sequencing (scRNA-seq) enables the resolution of cellular heterogeneity in diseases and facilitates the identification of novel cell types and subtypes. However, the grouping effects caused by cell-cell interactions are often overlooked in the development of tools for identifying subpopulations. We proposed LP_SGL which incorporates cell group structure to identify phenotype-associated subpopulations by integrating scRNA-seq, bulk expression and bulk phenotype data. Cell groups from scRNA-seq data were obtained by the Leiden algorithm, which facilitates the identification of subpopulations and improves model robustness. LP_SGL identified a higher percentage of cancer cells, T cells and tumor-associated cells than Scissor and scAB on lung adenocarcinoma diagnosis, melanoma drug response and liver cancer survival datasets, respectively. Biological analysis on three original datasets and four independent external validation sets demonstrated that the signaling genes of this cell subset can predict cancer, immunotherapy and survival.
Keywords: biological analysis; cell subpopulation; cell–cell interaction; data integration.
© The Author(s) 2023. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

References
-
- Suvà Mario L, Tirosh I. Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol Cell 2019;75(1):7–12. - PubMed
-
- Zhao J, Zhao B, Song X, et al. Subtype-DCC: decoupled contrastive clustering method for cancer subtype identification based on multi-omics data. Brief Bioinform 2023;24(2): bbad025. - PubMed
-
- Kaushik AC, Wang YJ, Wang X, Wei DQ. Irinotecan and vandetanib create synergies for treatment of pancreatic cancer patients with concomitant TP53 and KRAS mutations. Brief Bioinform 2021;22(3): bbaa149. - PubMed
-
- Yofe I, Dahan R, Amit I. Single-cell genomic approaches for developing the next generation of immunotherapies. Nat Med 2020;26(2):171–7. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

