Unveiling human origins of replication using deep learning: accurate prediction and comprehensive analysis
- PMID: 38008420
- PMCID: PMC10676776
- DOI: 10.1093/bib/bbad432
Unveiling human origins of replication using deep learning: accurate prediction and comprehensive analysis
Abstract
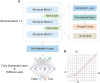
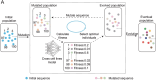
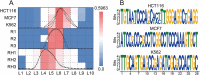
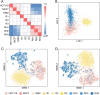
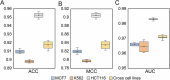
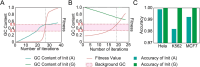
Accurate identification of replication origins (ORIs) is crucial for a comprehensive investigation into the progression of human cell growth and cancer therapy. Here, we proposed a computational approach Ori-FinderH, which can efficiently and precisely predict the human ORIs of various lengths by combining the Z-curve method with deep learning approach. Compared with existing methods, Ori-FinderH exhibits superior performance, achieving an area under the receiver operating characteristic curve (AUC) of 0.9616 for K562 cell line in 10-fold cross-validation. In addition, we also established a cross-cell-line predictive model, which yielded a further improved AUC of 0.9706. The model was subsequently employed as a fitness function to support genetic algorithm for generating artificial ORIs. Sequence analysis through iORI-Euk revealed that a vast majority of the created sequences, specifically 98% or more, incorporate at least one ORI for three cell lines (Hela, MCF7 and K562). This innovative approach could provide more efficient, accurate and comprehensive information for experimental investigation, thereby further advancing the development of this field.
Keywords: Z-curve method; deep learning; human genome; origin of replication.
© The Author(s) 2023. Published by Oxford University Press.
Figures

Similar articles
-
A computational platform to identify origins of replication sites in eukaryotes.Brief Bioinform. 2021 Mar 22;22(2):1940-1950. doi: 10.1093/bib/bbaa017. Brief Bioinform. 2021. PMID: 32065211
-
PLANNER: a multi-scale deep language model for the origins of replication site prediction.IEEE J Biomed Health Inform. 2024 Jan 4;PP. doi: 10.1109/JBHI.2024.3349584. Online ahead of print. IEEE J Biomed Health Inform. 2024. PMID: 38190667
-
A deep learning framework combined with word embedding to identify DNA replication origins.Sci Rep. 2021 Jan 12;11(1):844. doi: 10.1038/s41598-020-80670-x. Sci Rep. 2021. PMID: 33436981 Free PMC article.
-
Recent Advances on the Machine Learning Methods in Identifying DNA Replication Origins in Eukaryotic Genomics.Front Genet. 2018 Dec 10;9:613. doi: 10.3389/fgene.2018.00613. eCollection 2018. Front Genet. 2018. PMID: 30619452 Free PMC article. Review.
-
Recent development of Ori-Finder system and DoriC database for microbial replication origins.Brief Bioinform. 2019 Jul 19;20(4):1114-1124. doi: 10.1093/bib/bbx174. Brief Bioinform. 2019. PMID: 29329409 Review.
Cited by
-
Nmix: a hybrid deep learning model for precise prediction of 2'-O-methylation sites based on multi-feature fusion and ensemble learning.Brief Bioinform. 2024 Sep 23;25(6):bbae601. doi: 10.1093/bib/bbae601. Brief Bioinform. 2024. PMID: 39550226 Free PMC article.
-
DeOri 10.0: An Updated Database of Experimentally Identified Eukaryotic Replication Origins.Genomics Proteomics Bioinformatics. 2024 Dec 3;22(5):qzae076. doi: 10.1093/gpbjnl/qzae076. Genomics Proteomics Bioinformatics. 2024. PMID: 39404857 Free PMC article.
-
DNA sequence analysis landscape: a comprehensive review of DNA sequence analysis task types, databases, datasets, word embedding methods, and language models.Front Med (Lausanne). 2025 Apr 8;12:1503229. doi: 10.3389/fmed.2025.1503229. eCollection 2025. Front Med (Lausanne). 2025. PMID: 40265190 Free PMC article. Review.
References
-
- Bleichert F, Botchan MR, Berger JM. Mechanisms for initiating cellular DNA replication. Science 2017;355:eaah6317. - PubMed

