Targeted sequencing of high-density SNPs provides an enhanced tool for forensic applications and genetic landscape exploration in Chinese Korean ethnic group
- PMID: 38008719
- PMCID: PMC10680316
- DOI: 10.1186/s40246-023-00541-0
Targeted sequencing of high-density SNPs provides an enhanced tool for forensic applications and genetic landscape exploration in Chinese Korean ethnic group
Abstract
Background: In this study, we present a NGS-based panel designed for sequencing 1993 SNP loci for forensic DNA investigation. This panel addresses unique challenges encountered in forensic practice and allows for a comprehensive population genetic study of the Chinese Korean ethnic group. To achieve this, we combine our results with datasets from the 1000 Genomes Project and the Human Genome Diversity Panel.
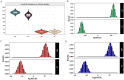
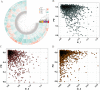
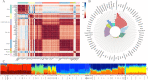
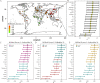
Results: We demonstrate that this panel is a reliable tool for individual identification and parentage testing, even when dealing with degraded DNA samples featuring exceedingly low SNP detection rates. The performance of this panel for complex kinship determinations, such as half-sibling and grandparent-grandchild scenarios, is also validated by various kinship simulations. Population genetic studies indicate that this panel can uncover population substructures on both global and regional scales. Notably, the Han population can be distinguished from the ethnic minorities in the northern and southern regions of East Asia, suggesting its potential for regional ancestry inference. Furthermore, we highlight that the Chinese Korean ethnic group, along with various Han populations from different regional areas and certain northern ethnic minorities (Daur, Tujia, Japanese, Mongolian, Xibo), exhibit a higher degree of genetic affinities when examined from a genomic perspective.
Conclusion: This study provides convincing evidence that the NGS-based panel can serve as a reliable tool for various forensic applications. Moreover, it has helped to enhance our knowledge about the genetic landscape of the Chinese Korean ethnic group.
Keywords: Chinese Korean ethnic group; Forensic application; Genetic structure; Next-generation sequencing; Single nucleotide polymorphism.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Oostdik K, Lenz K, Nye J, Schelling K, Yet D, Bruski S, Strong J, Buchanan C, Sutton J, Linner J, et al. Developmental validation of the PowerPlex(®) fusion system for analysis of casework and reference samples: a 24-locus multiplex for new database standards. Forensic Sci Int Genet. 2014;12:69–76. doi: 10.1016/j.fsigen.2014.04.013. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

