TRIM22 facilitates autophagosome-lysosome fusion by mediating the association of GABARAPs and PLEKHM1
- PMID: 38009729
- PMCID: PMC11135824
- DOI: 10.1080/15548627.2023.2287925
TRIM22 facilitates autophagosome-lysosome fusion by mediating the association of GABARAPs and PLEKHM1
Abstract
Tripartite motif (TRIM) proteins are a large family of E3 ubiquitin ligases implicated in antiviral defense systems, tumorigenesis, and protein quality control. TRIM proteins contribute to protein quality control by regulating the ubiquitin-proteasome system, endoplasmic reticulum-associated degradation, and macroautophagy/autophagy. However, the detailed mechanisms through which various TRIM proteins regulate downstream events have not yet been fully elucidated. Herein, we identified a novel function of TRIM22 in the regulation of autophagy. TRIM22 promotes autophagosome-lysosome fusion by mediating the association of GABARAP family proteins with PLEKHM1, thereby inducing the autophagic clearance of protein aggregates, independent of its E3 ubiquitin ligase activity. Furthermore, a TRIM22 variant associated with early-onset familial Alzheimer disease interferes with autophagosome-lysosome fusion and autophagic clearance. These findings suggest TRIM22 as a critical autophagic regulator that orchestrates autophagosome-lysosome fusion by scaffolding autophagy-related proteins, thus representing a potential therapeutic target in neurodegenerative diseases.Abbreviations: AD: Alzheimer disease; ADAOO: AD age of onset; AICD: APP intracellular domain; APP: amyloid beta precursor protein; BSA: bovine serum albumin; cDNAs: complementary DNAs; CQ: chloroquine; CTF: carboxyl-terminal fragment; EBSS: Earle's balanced salt solution; GABARAP: GABA type A receptor-associated protein; GST: glutathione S-transferase; HA: hemagglutinin; HOPS: homotypic fusion and protein sorting; IFN: interferon; IL1A/IL-1α: interleukin 1 alpha; KO: knockout; MTORC1: mechanistic target of rapamycin kinase complex 1; NFKBIA/IκBα: NFKB inhibitor alpha; NFE2L2/NRF2: NFE2 like bZIP transcription factor; PBS: phosphate-buffered saline; PI3K: class I phosphoinositide 3-kinase; PLA: proximity ligation assay; PLEKHM1: pleckstrin homology and RUN domain containing M1; PSEN1: presenilin 1; SEM: standard errors of the means; SNAREs: soluble N-ethylmaleimide-sensitive factor attachment protein receptors; SNCA: synuclein alpha; SNP: single nucleotide polymorphism; TBS: tris-buffered saline; TNF/TNF-α: tumor necrosis factor; TRIM: tripartite motif; ULK1: unc-51 like autophagy activating kinase 1; WT: wild-type.
Keywords: Alzheimer disease; PLEKHM1; TRIM22; autophagosome-lysosome fusion; autophagy.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures



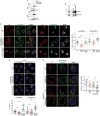

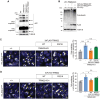


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
