Programmed chromosome fragmentation in ciliated protozoa: multiple means to chromosome ends
- PMID: 38009915
- PMCID: PMC10732028
- DOI: 10.1128/mmbr.00184-22
Programmed chromosome fragmentation in ciliated protozoa: multiple means to chromosome ends
Abstract
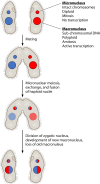
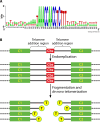
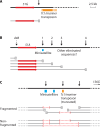
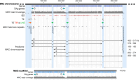
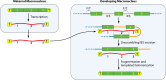
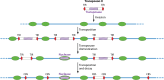
SUMMARYCiliated protozoa undergo large-scale developmental rearrangement of their somatic genomes when forming a new transcriptionally active macronucleus during conjugation. This process includes the fragmentation of chromosomes derived from the germline, coupled with the efficient healing of the broken ends by de novo telomere addition. Here, we review what is known of developmental chromosome fragmentation in ciliates that have been well-studied at the molecular level (Tetrahymena, Paramecium, Euplotes, Stylonychia, and Oxytricha). These organisms differ substantially in the fidelity and precision of their fragmentation systems, as well as in the presence or absence of well-defined sequence elements that direct excision, suggesting that chromosome fragmentation systems have evolved multiple times and/or have been significantly altered during ciliate evolution. We propose a two-stage model for the evolution of the current ciliate systems, with both stages involving repetitive or transposable elements in the genome. The ancestral form of chromosome fragmentation is proposed to have been derived from the ciliate small RNA/chromatin modification process that removes transposons and other repetitive elements from the macronuclear genome during development. The evolution of this ancestral system is suggested to have potentiated its replacement in some ciliate lineages by subsequent fragmentation systems derived from mobile genetic elements.
Keywords: IESs; Paramecium; Tetrahymena; chromosome fragmentation; ciliated protozoa; spirotrich; telomeres; transposons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Consensus inverted terminal repeat sequence of Paramecium IESs: resemblance to termini of Tc1-related and Euplotes Tec transposons.Nucleic Acids Res. 1995 Jun 11;23(11):2006-13. doi: 10.1093/nar/23.11.2006. Nucleic Acids Res. 1995. PMID: 7596830 Free PMC article.
-
The conjugation-specific Die5 protein is required for development of the somatic nucleus in both Paramecium and Tetrahymena.Eukaryot Cell. 2010 Jul;9(7):1087-99. doi: 10.1128/EC.00379-09. Epub 2010 May 21. Eukaryot Cell. 2010. PMID: 20495055 Free PMC article.
-
Transposon domestication versus mutualism in ciliate genome rearrangements.PLoS Genet. 2013;9(8):e1003659. doi: 10.1371/journal.pgen.1003659. Epub 2013 Aug 1. PLoS Genet. 2013. PMID: 23935529 Free PMC article. Review.
-
Internal eliminated sequences interrupting the Oxytricha 81 locus: allelic divergence, conservation, conversions, and possible transposon origins.Mol Biol Evol. 1996 Dec;13(10):1351-62. doi: 10.1093/oxfordjournals.molbev.a025581. Mol Biol Evol. 1996. PMID: 8952079
-
Developmentally programmed excision of internal DNA sequences in Paramecium aurelia.Biochimie. 2001 Nov-Dec;83(11-12):1009-22. doi: 10.1016/s0300-9084(01)01349-9. Biochimie. 2001. PMID: 11879729 Review.
Cited by
-
End resection and telomere healing of DNA double-strand breaks during nematode programmed DNA elimination.Nucleic Acids Res. 2024 Aug 27;52(15):8913-8929. doi: 10.1093/nar/gkae579. Nucleic Acids Res. 2024. PMID: 38953168 Free PMC article.
-
Genome content in the non-model ciliate Chilodonella uncinata: insights into nuclear architecture, gene-sized chromosomes among the total DNA in their somatic macronuclei during their development.bioRxiv [Preprint]. 2024 Nov 14:2024.11.13.623465. doi: 10.1101/2024.11.13.623465. bioRxiv. 2024. Update in: mSphere. 2025 Jun 25;10(6):e0007525. doi: 10.1128/msphere.00075-25. PMID: 39605396 Free PMC article. Updated. Preprint.
-
End resection and telomere healing of DNA double-strand breaks during nematode programmed DNA elimination.bioRxiv [Preprint]. 2024 Mar 16:2024.03.15.585292. doi: 10.1101/2024.03.15.585292. bioRxiv. 2024. Update in: Nucleic Acids Res. 2024 Aug 27;52(15):8913-8929. doi: 10.1093/nar/gkae579. PMID: 38559121 Free PMC article. Updated. Preprint.
-
Genome content reorganization in the non-model ciliate Chilodonella uncinata: insights into nuclear architecture, DNA content, and chromosome fragmentation during macronuclear development.mSphere. 2025 Jun 25;10(6):e0007525. doi: 10.1128/msphere.00075-25. Epub 2025 May 9. mSphere. 2025. PMID: 40340440 Free PMC article.
-
How and when organisms edit their own genomes.Nat Genet. 2025 Aug;57(8):1823-1834. doi: 10.1038/s41588-025-02230-1. Epub 2025 Jun 27. Nat Genet. 2025. PMID: 40579538 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

