The role of lysosomes as intermediates in betacoronavirus PHEV egress from nerve cells
- PMID: 38009916
- PMCID: PMC10734498
- DOI: 10.1128/jvi.01338-23
The role of lysosomes as intermediates in betacoronavirus PHEV egress from nerve cells
Abstract
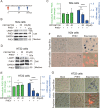
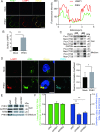
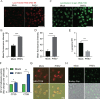
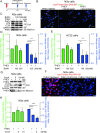
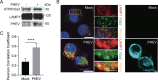
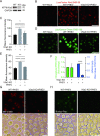
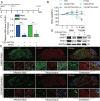
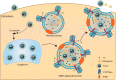
Betacoronaviruses, including severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and mouse hepatitis virus (MHV), exploit the lysosomal exocytosis pathway for egress. However, whether all betacoronaviruses members use the same pathway to exit cells remains unknown. Here, we demonstrated that porcine hemagglutinating encephalomyelitis virus (PHEV) egress occurs by Arl8b-dependent lysosomal exocytosis, a cellular egress mechanism shared by SARS-CoV-2 and MHV. Notably, PHEV acidifies lysosomes and activates lysosomal degradative enzymes, while SARS-CoV-2 and MHV deacidify lysosomes and limit the activation of lysosomal degradative enzymes. In addition, PHEV release depends on V-ATPase-mediated lysosomal pH. Furthermore, this is the first study to evaluate βCoV using lysosome for spreading through the body, and we have found that lysosome played a critical role in PHEV neural transmission and brain damage caused by virus infection in the central nervous system. Taken together, different betacoronaviruses could disrupt lysosomal function differently to exit cells.
Keywords: CNS; PHEV; V-ATPase; betacoronaviruses; lysosome; virus egress.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Vijgen L, Keyaerts E, Lemey P, Maes P, Van Reeth K, Nauwynck H, Pensaert M, Van Ranst M. 2006. Evolutionary history of the closely related group 2 Coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine Coronavirus, and human Coronavirus OC43. J Virol 80:7270–7274. doi:10.1128/JVI.02675-05 - DOI - PMC - PubMed
-
- Lan Y, Li Z, Wang Z, Wang X, Wang G, Zhang J, Hu S, Zhao K, Xu B, Gao F, He W. 2020. An experimental model of neurodegenerative disease based on porcine hemagglutinating encephalomyelitis virus-related lysosomal abnormalities. Mol Neurobiol 57:5299–5306. doi:10.1007/s12035-020-02105-y - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

