DNA hydroxymethylation differences underlie phenotypic divergence of somatic growth in Nile tilapia reared in common garden
- PMID: 38010265
- PMCID: PMC10732659
- DOI: 10.1080/15592294.2023.2282323
DNA hydroxymethylation differences underlie phenotypic divergence of somatic growth in Nile tilapia reared in common garden
Abstract
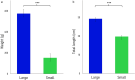
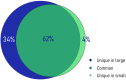
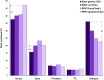
Phenotypic plasticity of metabolism and growth are essential for adaptation to new environmental conditions, such as those experienced during domestication. Epigenetic regulation plays a key role in this process but the underlying mechanisms are poorly understood, especially in the case of hydroxymethylation. Using reduced representation 5-hydroxymethylcytosine profiling, we compared the liver hydroxymethylomes in full-sib Nile tilapia with distinct growth rates (3.8-fold difference) and demonstrated that DNA hydroxymethylation is strongly associated with phenotypic divergence of somatic growth during the early stages of domestication. The 2677 differentially hydroxymethylated cytosines between fast- and slow-growing fish were enriched within gene bodies (79%), indicating a pertinent role in transcriptional regulation. Moreover, they were found in genes involved in biological processes related to skeletal system and muscle structure development, and there was a positive association between somatic growth and 5hmC levels in genes coding for growth factors, kinases and receptors linked to myogenesis. Single nucleotide polymorphism analysis revealed no genetic differentiation between fast- and slow-growing fish. In addition to unveiling a new link between DNA hydroxymethylation and epigenetic regulation of growth in fish during the initial stages of domestication, this study suggests that epimarkers may be applied in selective breeding programmes for superior phenotypes.
Keywords: DNA hydroxymethylation; Epigenetics; domestication; phenotypic plasticity; somatic growth; teleosts.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
