Nrf1 is not a direct target gene of SREBP1, albeit both are integrated into the rapamycin-responsive regulatory network in human hepatoma cells
- PMID: 38011090
- PMCID: PMC10681226
- DOI: 10.1371/journal.pone.0294508
Nrf1 is not a direct target gene of SREBP1, albeit both are integrated into the rapamycin-responsive regulatory network in human hepatoma cells
Abstract
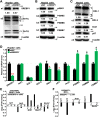
The essential role of protein degradation by ubiquitin-proteasome system is exerted primarily for maintaining cellular protein homeostasis. The transcriptional activation of proteasomal genes by mTORC1 signaling depends on Nrf1, but whether this process is directly via SREBP1 remains elusive. In this study, our experiment evidence revealed that Nrf1 is not a direct target of SREBP1, although both are involved in the rapamycin-responsive regulatory networks. Closely scrutinizing two distinct transcriptomic datasets unraveled no significant changes in transcriptional expression of Nrf1 and almost all proteasomal subunits in either siSREBP2-silencing cells or SREBP1-∕-MEFs, when compared to equivalent controls. However, distinct upstream signaling to Nrf1 dislocation by p97 and its processing by DDI1/2, along with downstream proteasomal expression, may be monitored by mTOR signaling, to various certain extents, depending on distinct experimental settings in different types of cells. Our further evidence has been obtained from DDI1-∕-(DDI2insC) cells, demonstrating that putative effects of mTOR on the rapamycin-responsive signaling to Nrf1 and proteasomes may also be executed partially through a DDI1/2-independent mechanism, albeit the detailed regulatory events remain to be determined.
Copyright: © 2023 Liu et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

