PARP-1 is a transcriptional rheostat of metabolic and bivalent genes during development
- PMID: 38012002
- PMCID: PMC10682175
- DOI: 10.26508/lsa.202302369
PARP-1 is a transcriptional rheostat of metabolic and bivalent genes during development
Abstract
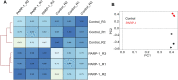
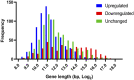
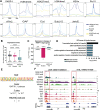
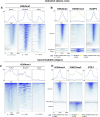
PARP-1 participates in various cellular processes, including gene regulation. In Drosophila, PARP-1 mutants undergo developmental arrest during larval-to-pupal transition. In this study, we investigated PARP-1 binding and its transcriptional regulatory role at this stage. Our findings revealed that PARP-1 binds and represses active metabolic genes, including glycolytic genes, whereas activating low-expression developmental genes, including a subset of "bivalent" genes in third-instar larvae. These bivalent promoters, characterized by dual enrichment of low H3K4me3 and high H3K27me3, a unimodal H3K4me1 enrichment at the transcription start site (conserved in C. elegans and zebrafish), H2Av depletion, and high accessibility, may persist throughout development. In PARP-1 mutant third-instar larvae, metabolic genes typically down-regulated during the larval-to-pupal transition in response to reduced energy needs were repressed by PARP-1. Simultaneously, developmental and bivalent genes typically active at this stage were activated by PARP-1. In addition, glucose and ATP levels were significantly reduced in PARP-1 mutants, suggesting an imbalance in metabolic regulation. We propose that PARP-1 is essential for maintaining the delicate balance between metabolic and developmental gene expression programs to ensure proper developmental progression.
© 2023 Bamgbose and Tulin.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
