scDREAMER for atlas-level integration of single-cell datasets using deep generative model paired with adversarial classifier
- PMID: 38012145
- PMCID: PMC10682386
- DOI: 10.1038/s41467-023-43590-8
scDREAMER for atlas-level integration of single-cell datasets using deep generative model paired with adversarial classifier
Abstract
Integration of heterogeneous single-cell sequencing datasets generated across multiple tissue locations, time, and conditions is essential for a comprehensive understanding of the cellular states and expression programs underlying complex biological systems. Here, we present scDREAMER ( https://github.com/Zafar-Lab/scDREAMER ), a data-integration framework that employs deep generative models and adversarial training for both unsupervised and supervised (scDREAMER-Sup) integration of multiple batches. Using six real benchmarking datasets, we demonstrate that scDREAMER can overcome critical challenges including skewed cell type distribution among batches, nested batch-effects, large number of batches and conservation of development trajectory across batches. Our experiments also show that scDREAMER and scDREAMER-Sup outperform state-of-the-art unsupervised and supervised integration methods respectively in batch-correction and conservation of biological variation. Using a 1 million cells dataset, we demonstrate that scDREAMER is scalable and can perform atlas-level cross-species (e.g., human and mouse) integration while being faster than other deep-learning-based methods.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
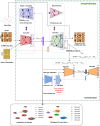
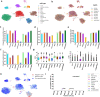


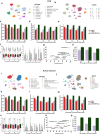
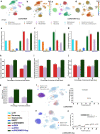
References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- figshare/10.6084/m9.figshare.24354295
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

