Mutual modulation of gut microbiota and the immune system in type 1 diabetes models
- PMID: 38012160
- PMCID: PMC10682479
- DOI: 10.1038/s41467-023-43652-x
Mutual modulation of gut microbiota and the immune system in type 1 diabetes models
Abstract
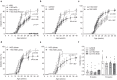
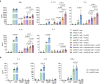
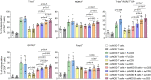
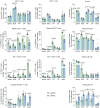
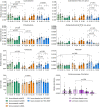
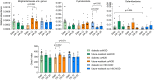
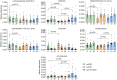
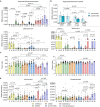
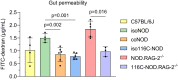
The transgenic 116C-NOD mouse strain exhibits a prevalent Th17 phenotype, and reduced type 1 diabetes (T1D) compared to non-obese diabetic (NOD) mice. A cohousing experiment between both models revealed lower T1D incidence in NOD mice cohoused with 116C-NOD, associated with gut microbiota changes, reduced intestinal permeability, shifts in T and B cell subsets, and a transition from Th1 to Th17 responses. Distinct gut bacterial signatures were linked to T1D in each group. Using a RAG-2-/- genetic background, we found that T cell alterations promoted segmented filamentous bacteria proliferation in young NOD and 116C-NOD, as well as in immunodeficient NOD.RAG-2-/- and 116C-NOD.RAG-2-/- mice across all ages. Bifidobacterium colonization depended on lymphocytes and thrived in a non-diabetogenic environment. Additionally, 116C-NOD B cells in 116C-NOD.RAG-2-/- mice enriched the gut microbiota in Adlercreutzia and reduced intestinal permeability. Collectively, these results indicate reciprocal modulation between gut microbiota and the immune system in rodent T1D models.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

