H3K36 methyltransferase NSD1 protects against osteoarthritis through regulating chondrocyte differentiation and cartilage homeostasis
- PMID: 38012390
- PMCID: PMC10781997
- DOI: 10.1038/s41418-023-01244-8
H3K36 methyltransferase NSD1 protects against osteoarthritis through regulating chondrocyte differentiation and cartilage homeostasis
Abstract
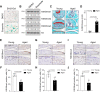
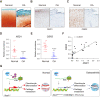
Osteoarthritis (OA) is one of the most common joint diseases, there are no effective disease-modifying drugs, and the pathological mechanisms of OA need further investigation. Here, we show that H3K36 methylations were decreased in senescent chondrocytes and age-related osteoarthritic cartilage. Prrx1-Cre inducible H3.3K36M transgenic mice showed articular cartilage destruction and osteophyte formation. Conditional knockout Nsd1Prrx1-Cre mice, but not Nsd2Prrx1-Cre or Setd2Prrx1-Cre mice, replicated the phenotype of K36M/+; Prrx1-Cre mice. Immunostaining results showed decreased anabolic and increased catabolic activities in Nsd1Prrx1-Cre mice, along with decreased chondrogenic differentiation. Transcriptome and ChIP-seq data revealed that Osr2 was a key factor affected by Nsd1. Intra-articular delivery of Osr2 adenovirus effectively improved the homeostasis of articular cartilage in Nsd1Prrx1-Cre mice. In human osteoarthritic cartilages, both mRNA and protein levels of NSD1 and OSR2 were decreased. Our results indicate that NSD1-induced H3K36 methylations and OSR2 expression play important roles in articular cartilage homeostasis and OA. Targeting H3K36 methylation and OSR2 would be a novel strategy for OA treatment.
© 2023. The Author(s), under exclusive licence to ADMC Associazione Differenziamento e Morte Cellulare.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
- 81672119/National Natural Science Foundation of China (National Science Foundation of China)
- 81725010/National Natural Science Foundation of China (National Science Foundation of China)
- 81902212/National Natural Science Foundation of China (National Science Foundation of China)
- 82202742/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

