AptaTrans: a deep neural network for predicting aptamer-protein interaction using pretrained encoders
- PMID: 38012571
- PMCID: PMC10680337
- DOI: 10.1186/s12859-023-05577-6
AptaTrans: a deep neural network for predicting aptamer-protein interaction using pretrained encoders
Abstract
Background: Aptamers, which are biomaterials comprised of single-stranded DNA/RNA that form tertiary structures, have significant potential as next-generation materials, particularly for drug discovery. The systematic evolution of ligands by exponential enrichment (SELEX) method is a critical in vitro technique employed to identify aptamers that bind specifically to target proteins. While advanced SELEX-based methods such as Cell- and HT-SELEX are available, they often encounter issues such as extended time consumption and suboptimal accuracy. Several In silico aptamer discovery methods have been proposed to address these challenges. These methods are specifically designed to predict aptamer-protein interaction (API) using benchmark datasets. However, these methods often fail to consider the physicochemical interactions between aptamers and proteins within tertiary structures.
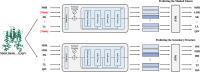
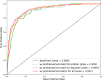
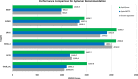
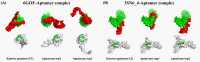
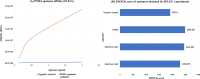
Results: In this study, we propose AptaTrans, a pipeline for predicting API using deep learning techniques. AptaTrans uses transformer-based encoders to handle aptamer and protein sequences at the monomer level. Furthermore, pretrained encoders are utilized for the structural representation. After validation with a benchmark dataset, AptaTrans has been integrated into a comprehensive toolset. This pipeline synergistically combines with Apta-MCTS, a generative algorithm for recommending aptamer candidates.
Conclusion: The results show that AptaTrans outperforms existing models for predicting API, and the efficacy of the AptaTrans pipeline has been confirmed through various experimental tools. We expect AptaTrans will enhance the cost-effectiveness and efficiency of SELEX in drug discovery. The source code and benchmark dataset for AptaTrans are available at https://github.com/pnumlb/AptaTrans .
Keywords: Aptamer protein interaction; Pretraing; SELEX; Structural representation; Transformer.
© 2023. The Author(s).
Conflict of interest statement
J.L. and H.Y.K. are full-time employees of Nuclixbio, a biopharmaceutical company that develops cell-penetrating biologics targeting intracellular oncoproteins with proprietary nanocarriers. This commercial affiliation does not alter our adherence to the
Figures
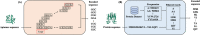







References
MeSH terms
Substances
Grants and funding
- 2021R1A2C2010775/National Research Foundation of Korea
- 2021R1A2C2010775/National Research Foundation of Korea
- 2021R1A2C2010775/National Research Foundation of Korea
- 2021R1A2C2010775/National Research Foundation of Korea
- 2021R1A2C2010775/National Research Foundation of Korea
- 2021R1A2C2010775/National Research Foundation of Korea
- IITP-2023-00254177/Institute of Information & Communications Technology Planning & Evaluation
- IITP-2023-00254177/Institute of Information & Communications Technology Planning & Evaluation
- IITP-2023-00254177/Institute of Information & Communications Technology Planning & Evaluation
- IITP-2023-00254177/Institute of Information & Communications Technology Planning & Evaluation
- IITP-2023-00254177/Institute of Information & Communications Technology Planning & Evaluation
LinkOut - more resources
Full Text Sources

