Research on radiotherapy related genes and prognostic target identification of rectal cancer based on multi-omics
- PMID: 38012642
- PMCID: PMC10680259
- DOI: 10.1186/s12967-023-04753-9
Research on radiotherapy related genes and prognostic target identification of rectal cancer based on multi-omics
Abstract
Background: Radiosensitivity of rectal cancer is related to the radiotherapy efficacy and prognosis of patients with rectal cancer, and the genes and molecular mechanisms related to radiosensitivity of rectal cancer have not been clarified. We explored the radiosensitivity related genes of rectal cancer at a multi omics level.
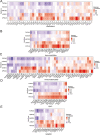
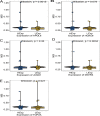
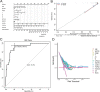
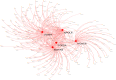
Methods: mRNA expression data and rectum adenocarcinoma (READ) data were obtained from the Cancer Genome Atlas (TCGA) and the Gene Expression Omnibus Database (GEO) (GSE150082, GSE60331, GSE46862, GSE46862). Differentially expressed genes between radiotherapy sensitive group and radiotherapy insensitive group were screened. GO analysis and KEGG pathway analysis were performed for differentially expressed genes. Among the differentially expressed genes, five core genes associated with rectal cancer prognosis were selected using random survival forest analysis. For these five core genes, drug sensitivity analysis, immune cell infiltration analysis, TISIDB database immune gene correlation analysis, GSEA enrichment analysis, construction of Nomogram prediction model, transcriptional regulatory network analysis, and qRT-PCR validation was performed on human rectal adenocarcinoma tissue.
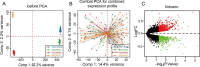
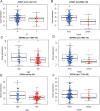
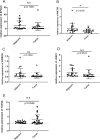
Results: We found that 600 up-regulated genes and 553 down-regulated genes were significantly different between radiotherapy sensitive group and radiotherapy insensitive group in rectal cancer. Five key genes, TOP2A, MATR3, APOL6, JOSD1, and HOXC6, were finally screened by random survival forest analysis. These five key genes were associated with different immune cell infiltration, immune-related genes, and chemosensitivity. A comprehensive transcriptional regulatory network was constructed based on these five core genes. qRT-PCR revealed that MATR3 expression was different in rectal cancer tissues and adjacent non-cancerous tissues, while APOL6, HOXC6, JOSD1, and TOP2A expression was not different.
Conclusion: Five radiosensitivity-related genes related to the prognosis of rectal cancer: TOP2A, MATR3, APOL6, JOSD1, HOXC6, are involved in multiple processes such as immune cell infiltration, immune-related genes, chemosensitivity, signaling pathways and transcriptional regulatory networks and may be potential biomarkers for radiotherapy of rectal cancer.
Keywords: Differentially expressed genes; Multi-omics; Prognosis; Radiosensitivity; Rectal cancer.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures








References
-
- Li YX, Wang LH, Gao L, et al. Oncology Radiotherapy. 5. China Union Medical University Press; 2018. pp. 1109–1125.
-
- Gambacorta MA, Masciocchi C, Chiloiro G, Meldolesi E, Macchia G, Vansoest J, Peters F, Collette L, Gerard JP, Ngan S, et al. Timing to achieve the highest rate of pCR after preoperative radiochemotherapy in rectal cancer: a pooled analysis of 3085 patients from 7 randomized trials. Radiother Oncol. 2021;154:154–160. doi: 10.1016/j.radonc.2020.09.026. - DOI - PubMed
-
- Kailiang W, Yan F, Chen XX, et al. Clinical Oncology Radiation Therapy. Fudan University Press; 2017. pp. 126–128.
-
- Chunyan H, Lijun S, Qingyun L. Mechanism and clinical research progress of radiotherapy combined with immunotherapy. Chin J Cancer Prev Treat. 2019;26(18):1317–1322.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

