Dysfunction of Drosophila mitochondrial carrier homolog (Mtch) alters apoptosis and disturbs development
- PMID: 38013241
- PMCID: PMC10839352
- DOI: 10.1002/2211-5463.13742
Dysfunction of Drosophila mitochondrial carrier homolog (Mtch) alters apoptosis and disturbs development
Abstract
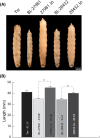
Mitochondrial carrier homologs 1 (MTCH1) and 2 (MTCH2) are orphan members of the mitochondrial transporter family SLC25. Human MTCH1 is also known as presenilin 1-associated protein, PSAP. MTCH2 is a receptor for tBid and is related to lipid metabolism. Both proteins have been recently described as protein insertases of the outer mitochondrial membrane. We have depleted Mtch in Drosophila and show here that mutant flies are unable to complete development, showing an excess of apoptosis during pupation; this observation was confirmed by RNAi in Schneider cells. These findings are contrary to what has been described in humans. We discuss the implications in view of recent reports concerning the function of these proteins.
Keywords: Drosophila; apoptosis; development; mitochondria; mitochondrial carrier homolog (MTCH).
© 2023 The Authors. FEBS Open Bio published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Comment on
-
The modified mitochondrial outer membrane carrier MTCH2 links mitochondrial fusion to lipogenesis.J Cell Biol. 2021 Nov 1;220(11):e202103122. doi: 10.1083/jcb.202103122. Epub 2021 Sep 29. J Cell Biol. 2021. PMID: 34586346 Free PMC article.
References
-
- Luo X, Budihardjo I, Zou H, Slaughter C and Wang X (1998) Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94, 481–490. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

