This is a preprint.
The MYC axis in advanced prostate cancer is impacted through concurrent targeting of ERβ and AR using a novel ERβ-selective ligand alongside Enzalutamide
- PMID: 38014010
- PMCID: PMC10680693
- DOI: 10.1101/2023.11.15.567282
The MYC axis in advanced prostate cancer is impacted through concurrent targeting of ERβ and AR using a novel ERβ-selective ligand alongside Enzalutamide
Abstract
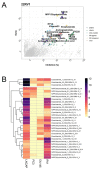
We have dissected the role of Estrogen receptor beta (ERβ) in prostate cancer (PCa) with a novel ERβ ligand, OSU-ERb-12. Drug screens revealed additive interactions between OSU-ERB-12 and either epigenetic inhibitors or the androgen receptor antagonist, Enzalutamide (Enza). Clonogenic and cell biolody studies supported the potent additive effects of OSU-ERB-12 (100nM) and Enza (1μM). The cooperative behavior was in PCa cell lines treated with either OSU-ERB-12 plus Enza or combinations involving 17β-estradiol (E2). OSU-ERb-12 plus Enza uniquely impacted the transcriptiome, accessible chromatin, and the AR, MYC and H3K27ac cistromes. This included skewed transcriptional responses including suppression of the androgen and MYC transcriptomes, and repressed MYC protein. OSU-ERb-12 plus Enza uniquely impacted chromatin accessibility at approximately 3000 nucleosome-free sites, enriched at enhancers, enriched for basic Helix-Loop-Helix motifs. CUT&RUN experiments revealed combination treatment targeting of MYC, AR, and H3K27ac again shaping enhancer accessibility. Specifically, it repressed MYC interactions at enhancer regions enriched for bHLH motifs, and overlapped with publicly-available bHLH cistromes. Finally, cistrome-transcriptome analyses identified ~200 genes that distinguished advanced PCa tumors in the SU2C cohort with high androgen and low neuroendocrine scores.
Conflict of interest statement
Competing interests CCC and CEB hold pending patents on methods of use for OSU-ERb-12. All other authors certify that he or she has NO affiliations with or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements), or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
Figures





References
-
- Davies A, Zoubeidi A, Beltran H, Selth LA. The Transcriptional and Epigenetic Landscape of Cancer Cell Lineage Plasticity. Cancer Discov 2023;13(8):1771–88 doi 10.1158/2159-8290.CD-23-0225. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
