This is a preprint.
Aberrant splicing exonizes C9ORF72 repeat expansion in ALS/FTD
- PMID: 38014069
- PMCID: PMC10680656
- DOI: 10.1101/2023.11.13.566896
Aberrant splicing exonizes C9ORF72 repeat expansion in ALS/FTD
Update in
-
Aberrant splicing exonizes C9orf72 repeat expansion in ALS/FTD.Nat Neurosci. 2025 Oct;28(10):2034-2043. doi: 10.1038/s41593-025-02039-5. Epub 2025 Aug 11. Nat Neurosci. 2025. PMID: 40790269 Free PMC article.
Abstract
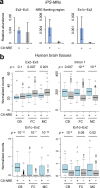
A nucleotide repeat expansion (NRE) in the first annotated intron of the C9ORF72 gene is the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). While C9 NRE-containing RNAs can be translated into several toxic dipeptide repeat proteins, how an intronic NRE can assess the translation machinery in the cytoplasm remains unclear. By capturing and sequencing NRE-containing RNAs from patient-derived cells, we found that C9 NRE was exonized by the usage of downstream 5' splice sites and exported from the nucleus in a variety of spliced mRNA isoforms. C9ORF72 aberrant splicing was substantially elevated in both C9 NRE+ motor neurons and human brain tissues. Furthermore, NREs above the pathological threshold were sufficient to activate cryptic splice sites in reporter mRNAs. In summary, our results revealed a crucial and potentially widespread role of repeat-induced aberrant splicing in the biogenesis, localization, and translation of NRE-containing RNAs.
Keywords: ALS/FTD; C9ORF72; cryptic splice site; mRNA splicing; nucleotide repeat expansion.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
