This is a preprint.
Network Enrichment Significance Testing in Brain-Phenotype Association Studies
- PMID: 38014137
- PMCID: PMC10680593
- DOI: 10.1101/2023.11.10.566593
Network Enrichment Significance Testing in Brain-Phenotype Association Studies
Update in
-
Network enrichment significance testing in brain-phenotype association studies.Hum Brain Mapp. 2024 Jun 1;45(8):e26714. doi: 10.1002/hbm.26714. Hum Brain Mapp. 2024. PMID: 38878300 Free PMC article.
Abstract
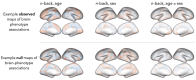
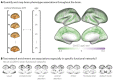
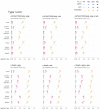
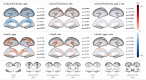
Functional networks often guide our interpretation of spatial maps of brain-phenotype associations. However, methods for assessing enrichment of associations within networks of interest have varied in terms of both scientific rigor and underlying assumptions. While some approaches have relied on subjective interpretations, others have made unrealistic assumptions about the spatial structure of imaging data, leading to inflated false positive rates. We seek to address this gap in existing methodology by borrowing insight from a method widely used in genomics research for testing enrichment of associations between a set of genes and a phenotype of interest. We propose Network Enrichment Significance Testing (NEST), a flexible framework for testing the specificity of brain-phenotype associations to functional networks or other sub-regions of the brain. We apply NEST to study phenotype associations with structural and functional brain imaging data from a large-scale neurodevelopmental cohort study.
Conflict of interest statement
Russell T. Shinohara receives consulting income from Octave Bioscience and compensation for reviewership duties from the American Medical Association. Aaron Alexander-Bloch receives consulting income from Octave Bioscience and holds equity and serves on the board of directors of Centile Biosciences. Mingyao Li receives research funding from Biogen Inc. that is unrelated to the current manuscript.
Figures


References
-
- Baller Erica B, Valcarcel Alessandra M, Adebimpe Azeez, Alexander-Bloch Aaron, Cui Zaixu, Gur Ruben C, Gur Raquel E, Larsen Bart L, Linn Kristin A, O’Donnell Carly M, et al. (2022). “Developmental coupling of cerebral blood flow and fMRI fluctuations in youth”. In: Cell Reports 38.13, p. 110576. - PMC - PubMed
-
- Baum Graham L, Cui Zaixu, Roalf David R, Ciric Rastko, Betzel Richard F, Larsen Bart, Cieslak Matthew, Cook Philip A, Xia Cedric H, Moore Tyler M, et al. (2020). “Development of structure–function coupling in human brain networks during youth”. In: Proceedings of the National Academy of Sciences 117.1, pp. 771–778. - PMC - PubMed
-
- Belzunce Felix, Riquelme Carolina Martinez, and Mulero Julio (2015). An introduction to stochastic orders. Academic Press.
-
- Burt Joshua B, Helmer Markus, Shinn Maxwell, Anticevic Alan, and Murray John D (2020). “Generative modeling of brain maps with spatial autocorrelation”. In: NeuroImage 220, p. 117038. - PubMed
Publication types
Grants and funding
- R37 MH125829/MH/NIMH NIH HHS/United States
- R01 MH120482/MH/NIMH NIH HHS/United States
- K23 MH133118/MH/NIMH NIH HHS/United States
- L30 MH127652/MH/NIMH NIH HHS/United States
- R01 MH132934/MH/NIMH NIH HHS/United States
- R01 MH120174/MH/NIMH NIH HHS/United States
- R01 MH119219/MH/NIMH NIH HHS/United States
- R01 MH119185/MH/NIMH NIH HHS/United States
- R01 MH112847/MH/NIMH NIH HHS/United States
- R01 EB022573/EB/NIBIB NIH HHS/United States
- R01 NS112274/NS/NINDS NIH HHS/United States
- R01 MH123550/MH/NIMH NIH HHS/United States
- R01 MH123563/MH/NIMH NIH HHS/United States
- R01 MH113550/MH/NIMH NIH HHS/United States
- ZIA MH002949/ImNIH/Intramural NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous
