This is a preprint.
Non-muscle myosin II regulates presynaptic actin assemblies and neuronal mechanobiology in Drosophila
- PMID: 38014140
- PMCID: PMC10680633
- DOI: 10.1101/2023.11.10.566609
Non-muscle myosin II regulates presynaptic actin assemblies and neuronal mechanobiology in Drosophila
Update in
-
Nonmuscle myosin II regulates presynaptic actin and neuronal mechanobiology in Drosophila.J Cell Biol. 2025 Sep 1;224(9):e202501211. doi: 10.1083/jcb.202501211. Epub 2025 Jul 11. J Cell Biol. 2025. PMID: 40644605
Abstract
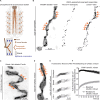
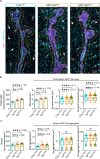
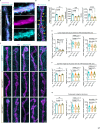
Neuromuscular junctions (NMJs) are evolutionarily ancient, specialized contacts between neurons and muscles. They endure mechanical strain from muscle contractions throughout life, but cellular mechanisms for managing this stress remain unclear. Here we identify a novel actomyosin structure at Drosophila larval NMJs, consisting of a long-lived, low-turnover presynaptic actin core that co-localizes with non-muscle myosin II (NMII). This core is likely to have contractile properties, as manipulating neuronal NMII levels or activity disrupts its organization. Intriguingly, depleting neuronal NMII triggered changes in postsynaptic muscle NMII levels and organization near synapses, suggesting transsynaptic propagation of actomyosin rearrangements. We also found reduced levels of Integrin adhesion receptors both pre- and postsynaptically upon NMII knockdown, indicating disrupted neuron-muscle connections. Mechanical severing of axons caused similar actin core fragmentation and Integrin loss to NMII depletion, suggesting this structure responds to tension. Our findings reveal a presynaptic actomyosin assembly that maintains mechanical continuity between neurons and muscle, possibly facilitating mechanotransduction at the NMJ via Integrin-mediated adhesion.
Keywords: Drosophila; actin cytoskeleton; integrin; mechanobiology; neuromuscular junction; non-muscle myosin II.
Conflict of interest statement
Competing interests: The authors declare no competing interests. J.B. has received ad hoc consultancy compensation for activities with Novartis, Sanofi, CSL Behring, Alnylam, Roche, ARGENX and Amylyx.
Figures





Similar articles
-
Nonmuscle myosin II regulates presynaptic actin and neuronal mechanobiology in Drosophila.J Cell Biol. 2025 Sep 1;224(9):e202501211. doi: 10.1083/jcb.202501211. Epub 2025 Jul 11. J Cell Biol. 2025. PMID: 40644605
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Rho/Rok-dependent regulation of actomyosin contractility at tricellular junctions restricts epithelial permeability in Drosophila.Curr Biol. 2025 Mar 24;35(6):1181-1196.e5. doi: 10.1016/j.cub.2025.01.043. Epub 2025 Feb 17. Curr Biol. 2025. PMID: 39965573
-
Omega-3 fatty acids for depression in adults.Cochrane Database Syst Rev. 2015 Nov 5;2015(11):CD004692. doi: 10.1002/14651858.CD004692.pub4. Cochrane Database Syst Rev. 2015. Update in: Cochrane Database Syst Rev. 2021 Nov 24;11:CD004692. doi: 10.1002/14651858.CD004692.pub5. PMID: 26537796 Free PMC article. Updated.
-
Corticosteroids for the treatment of Duchenne muscular dystrophy.Cochrane Database Syst Rev. 2016 May 5;2016(5):CD003725. doi: 10.1002/14651858.CD003725.pub4. Cochrane Database Syst Rev. 2016. PMID: 27149418 Free PMC article.
References
-
- Abouelezz A., Stefen H., Segerstråle M., Micinski D., Minkeviciene R., Lahti L., Hardeman E.C., Gunning P.W., Hoogenraad C.C., Taira T., Fath T., and Hotulainen P.. 2020. Tropomyosin Tpm3.1 Is Required to Maintain the Structure and Function of the Axon Initial Segment. iScience. 23:101053. - PMC - PubMed
-
- Arganda-Carreras I., Kaynig V., Rueden C., Eliceiri K.W., Schindelin J., Cardona A., and Sebastian Seung H.. 2017. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics. 33:2424–2426. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
