This is a preprint.
DeepN4: Learning N4ITK Bias Field Correction for T1-weighted Images
- PMID: 38014176
- PMCID: PMC10680935
- DOI: 10.21203/rs.3.rs-3585882/v1
DeepN4: Learning N4ITK Bias Field Correction for T1-weighted Images
Update in
-
DeepN4: Learning N4ITK Bias Field Correction for T1-weighted Images.Neuroinformatics. 2024 Apr;22(2):193-205. doi: 10.1007/s12021-024-09655-9. Epub 2024 Mar 25. Neuroinformatics. 2024. PMID: 38526701 Free PMC article.
Abstract
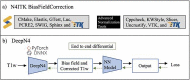
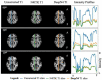
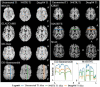
T1-weighted (T1w) MRI has low frequency intensity artifacts due to magnetic field inhomogeneities. Removal of these biases in T1w MRI images is a critical preprocessing step to ensure spatially consistent image interpretation. N4ITK bias field correction, the current state-of-the-art, is implemented in such a way that makes it difficult to port between different pipelines and workflows, thus making it hard to reimplement and reproduce results across local, cloud, and edge platforms. Moreover, N4ITK is opaque to optimization before and after its application, meaning that methodological development must work around the inhomogeneity correction step. Given the importance of bias fields correction in structural preprocessing and flexible implementation, we pursue a deep learning approximation / reinterpretation of the N4ITK bias fields correction to create a method which is portable, flexible, and fully differentiable. In this paper, we trained a deep learning network "DeepN4" on eight independent cohorts from 72 different scanners and age ranges with N4ITK-corrected T1w MRI and bias field for supervision in log space. We found that we can closely approximate N4ITK bias fields correction with naïve networks. We evaluate the peak signal to noise ratio (PSNR) in test dataset against the N4ITK corrected images. The median PSNR of corrected images between N4ITK and DeepN4 was 47.96 dB. In addition, we assess the DeepN4 model on eight additional external datasets and show the generalizability of the approach. This study establishes that incompatible N4ITK preprocessing steps can be closely approximated by naïve deep neural networks, facilitating more flexibility. All code and models are released at https://github.com/MASILab/DeepN4.
Keywords: 3D U-Net; N4ITK; T1-weighted images; bias field correction; inhomogeneity.
Conflict of interest statement
Conflict of Interest The authors declare that they have no conflict of interest
Figures


References
-
- Damadian R. Tumor detection by nuclear magnetic resonance. Science. 1971;171(3976):1151–1153. - PubMed
-
- Johnson K. Basic proton MR imaging: tissue signal characteristics. Harvard Medical School. 2016:230–231.
-
- Vovk U, Pernus F, Likar B. A review of methods for correction of intensity inhomogeneity in MRI. IEEE transactions on medical imaging. 2007;26(3):405–421. - PubMed
Publication types
Grants and funding
- R01 AG056534/AG/NIA NIH HHS/United States
- P30 AG066515/AG/NIA NIH HHS/United States
- P30 AG066530/AG/NIA NIH HHS/United States
- P30 AG072972/AG/NIA NIH HHS/United States
- R01 AG034962/AG/NIA NIH HHS/United States
- P30 AG072979/AG/NIA NIH HHS/United States
- K01 AG073584/AG/NIA NIH HHS/United States
- P20 AG068082/AG/NIA NIH HHS/United States
- P30 AG072975/AG/NIA NIH HHS/United States
- P30 AG066444/AG/NIA NIH HHS/United States
- R01 AG043434/AG/NIA NIH HHS/United States
- UL1 TR000445/TR/NCATS NIH HHS/United States
- P30 AG066507/AG/NIA NIH HHS/United States
- P30 AG072946/AG/NIA NIH HHS/United States
- P30 AG066518/AG/NIA NIH HHS/United States
- P20 AG068053/AG/NIA NIH HHS/United States
- R01 EB017230/EB/NIBIB NIH HHS/United States
- P30 AG066511/AG/NIA NIH HHS/United States
- U24 AG072122/AG/NIA NIH HHS/United States
- P30 AG066512/AG/NIA NIH HHS/United States
- P30 AG062421/AG/NIA NIH HHS/United States
- U01 AG024904/AG/NIA NIH HHS/United States
- P30 AG066508/AG/NIA NIH HHS/United States
- P01 AG003991/AG/NIA NIH HHS/United States
- K01 EB032898/EB/NIBIB NIH HHS/United States
- P30 AG072978/AG/NIA NIH HHS/United States
- P01 AG026276/AG/NIA NIH HHS/United States
- P30 AG062429/AG/NIA NIH HHS/United States
- P30 AG066519/AG/NIA NIH HHS/United States
- R01 AG062826/AG/NIA NIH HHS/United States
- U24 AG074855/AG/NIA NIH HHS/United States
- P30 AG072973/AG/NIA NIH HHS/United States
- P30 AG062422/AG/NIA NIH HHS/United States
- R01 AG079280/AG/NIA NIH HHS/United States
- R01 EB009352/EB/NIBIB NIH HHS/United States
- P30 AG066462/AG/NIA NIH HHS/United States
- P50 AG005133/AG/NIA NIH HHS/United States
- P30 AG066509/AG/NIA NIH HHS/United States
- U54 MH091657/MH/NIMH NIH HHS/United States
- T32 GM007347/GM/NIGMS NIH HHS/United States
- P20 AG068077/AG/NIA NIH HHS/United States
- S10 OD023680/OD/NIH HHS/United States
- P30 AG066546/AG/NIA NIH HHS/United States
- UL1 TR000448/TR/NCATS NIH HHS/United States
- P30 AG072977/AG/NIA NIH HHS/United States
- P30 AG062677/AG/NIA NIH HHS/United States
- P50 HD103537/HD/NICHD NIH HHS/United States
- P20 AG068024/AG/NIA NIH HHS/United States
- P30 AG072958/AG/NIA NIH HHS/United States
- P30 AG062715/AG/NIA NIH HHS/United States
- UL1 RR024975/RR/NCRR NIH HHS/United States
- P30 AG066506/AG/NIA NIH HHS/United States
- P30 AG066468/AG/NIA NIH HHS/United States
- P30 AG072976/AG/NIA NIH HHS/United States
- P30 AG072947/AG/NIA NIH HHS/United States
- P30 AG072931/AG/NIA NIH HHS/United States
- U01 AG033655/AG/NIA NIH HHS/United States
- P30 AG066514/AG/NIA NIH HHS/United States
- P30 AG072959/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

