This is a preprint.
Loss of cis-PTase function in the liver promotes a highly penetrant form of fatty liver disease that rapidly transitions to hepatocellular carcinoma
- PMID: 38014178
- PMCID: PMC10680637
- DOI: 10.1101/2023.11.13.566870
Loss of cis-PTase function in the liver promotes a highly penetrant form of fatty liver disease that rapidly transitions to hepatocellular carcinoma
Abstract
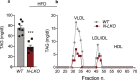
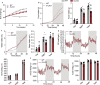
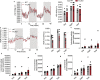
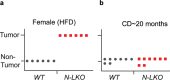
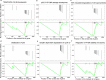
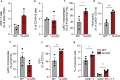
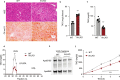
Obesity-linked fatty liver is a significant risk factor for hepatocellular carcinoma (HCC)1,2; however, the molecular mechanisms underlying the transition from non-alcoholic fatty liver disease (NAFLD) to HCC remains unclear. The present study explores the role of the endoplasmic reticulum (ER)-associated protein NgBR, an essential component of the cis-prenyltransferases (cis-PTase) enzyme3, in chronic liver disease. Here we show that genetic depletion of NgBR in hepatocytes of mice (N-LKO) intensifies triacylglycerol (TAG) accumulation, inflammatory responses, ER/oxidative stress, and liver fibrosis, ultimately resulting in HCC development with 100% penetrance after four months on a high-fat diet. Comprehensive genomic and single cell transcriptomic atlas from affected livers provides a detailed molecular analysis of the transition from liver pathophysiology to HCC development. Importantly, pharmacological inhibition of diacylglycerol acyltransferase-2 (DGAT2), a key enzyme in hepatic TAG synthesis, abrogates diet-induced liver damage and HCC burden in N-LKO mice. Overall, our findings establish NgBR/cis-PTase as a critical suppressor of NAFLD-HCC conversion and suggests that DGAT2 inhibition may serve as a promising therapeutic approach to delay HCC formation in patients with advanced non-alcoholic steatohepatitis (NASH).
Figures















References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
