This is a preprint.
Fast and accurate local ancestry inference with Recomb-Mix
- PMID: 38014185
- PMCID: PMC10680832
- DOI: 10.1101/2023.11.17.567650
Fast and accurate local ancestry inference with Recomb-Mix
Update in
-
Recomb-Mix: fast and accurate local ancestry inference.Bioinformatics. 2025 Jul 1;41(Supplement_1):i180-i188. doi: 10.1093/bioinformatics/btaf227. Bioinformatics. 2025. PMID: 40662780 Free PMC article.
Abstract
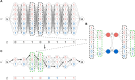
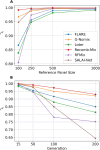
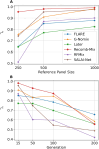
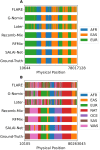
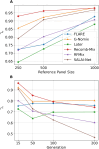
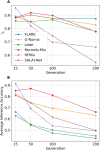
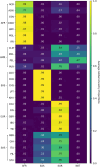
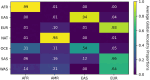
The availability of large genotyped cohorts brings new opportunities for revealing the high-resolution genetic structure of admixed populations via local ancestry inference (LAI), the process of identifying the ancestry of each segment of an individual haplotype. Though current methods achieve high accuracy in standard cases, LAI is still challenging when reference populations are more similar (e.g., intra-continental), when the number of reference populations is too numerous, or when the admixture events are deep in time, all of which are increasingly unavoidable in large biobanks. Here, we present a new LAI method, Recomb-Mix. Recomb-Mix integrates the elements of existing methods of the site-based Li and Stephens model and introduces a new graph collapsing trick to simplify counting paths with the same ancestry label readout. Through comprehensive benchmarking on various simulated datasets, we show that Recomb-Mix is more accurate than existing methods in diverse sets of scenarios while being competitive in terms of resource efficiency. We expect that Recomb-Mix will be a useful method for advancing genetics studies of admixed populations.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
