This is a preprint.
Spatial proteo-transcriptomic profiling reveals the molecular landscape of borderline ovarian tumors and their invasive progression
- PMID: 38014221
- PMCID: PMC10680885
- DOI: 10.1101/2023.11.13.23298409
Spatial proteo-transcriptomic profiling reveals the molecular landscape of borderline ovarian tumors and their invasive progression
Update in
-
Spatial proteo-transcriptomic profiling reveals the molecular landscape of borderline ovarian tumors and their invasive progression.Cancer Cell. 2025 Aug 11;43(8):1495-1511.e7. doi: 10.1016/j.ccell.2025.06.004. Epub 2025 Jun 26. Cancer Cell. 2025. PMID: 40578359
Abstract
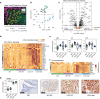
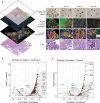
Serous borderline tumors (SBT) are epithelial neoplastic lesions of the ovaries that commonly have a good prognosis. In 10-15% of cases, however, SBT will recur as low-grade serous cancer (LGSC), which is deeply invasive and responds poorly to current standard chemotherapy1,2,3. While genetic alterations suggest a common origin, the transition from SBT to LGSC remains poorly understood4. Here, we integrate spatial proteomics5 with spatial transcriptomics to elucidate the evolution from SBT to LGSC and its corresponding metastasis at the molecular level in both the stroma and the tumor. We show that the transition of SBT to LGSC occurs in the epithelial compartment through an intermediary stage with micropapillary features (SBT-MP), which involves a gradual increase in MAPK signaling. A distinct subset of proteins and transcripts was associated with the transition to invasive tumor growth, including the neuronal splicing factor NOVA2, which was limited to expression in LGSC and its corresponding metastasis. An integrative pathway analysis exposed aberrant molecular signaling of tumor cells supported by alterations in angiogenesis and inflammation in the tumor microenvironment. Integration of spatial transcriptomics and proteomics followed by knockdown of the most altered genes or pharmaceutical inhibition of the most relevant targets confirmed their functional significance in regulating key features of invasiveness. Combining cell-type resolved spatial proteomics and transcriptomics allowed us to elucidate the sequence of tumorigenesis from SBT to LGSC. The approach presented here is a blueprint to systematically elucidate mechanisms of tumorigenesis and find novel treatment strategies.
Conflict of interest statement
Competing interests E.L. receives research funding to study ovarian cancer from Arsenal Bioscience and AbbVie through the University of Chicago unrelated to this work. The authors declare no competing interests in the context of this manuscript.
Figures




References
-
- Gershenson D. M. et al. Recurrent low-grade serous ovarian carcinoma is relatively chemoresistant. Gynecol Oncol 114, 48–52 (2009). - PubMed
-
- Tang M. et al. PARAGON: A Phase II study of anastrozole in patients with estrogen receptorpositive recurrent/metastatic low-grade ovarian cancers and serous borderline ovarian tumors. Gynecol. Oncol. 154, 531–538 (2019). - PubMed
-
- Singer G. et al. Patterns of p53 mutations separate ovarian serous borderline tumors and low- and high-grade carcinomas and provide support for a new model of ovarian carcinogenesis: A mutational analysis with immunohistochemical correlation. American Journal of Surgical Pathology 29, 218–224 (2005). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
