This is a preprint.
Electronic Polarizability Tunes the Function of the Human Bestrophin 1 Cl- Channel
- PMID: 38014257
- PMCID: PMC10680768
- DOI: 10.1101/2023.11.14.567055
Electronic Polarizability Tunes the Function of the Human Bestrophin 1 Cl- Channel
Update in
-
Electronic Polarizability Tunes the Function of the Human Bestrophin 1 Cl- Channel.J Chem Theory Comput. 2025 Jan 28;21(2):933-942. doi: 10.1021/acs.jctc.4c01039. Epub 2025 Jan 3. J Chem Theory Comput. 2025. PMID: 39754290 Free PMC article.
Abstract
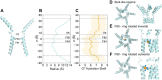
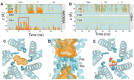
Mechanisms of anion permeation within ion channels and nanopores remain poorly understood. Recent cryo-electron microscopy structures of the human bestrophin 1 Cl- channel (hBest1) provide an opportunity to evaluate ion interactions predicted by molecular dynamics (MD) simulations against experimental observations. Here, we implement the fully polarizable forcefield AMOEBA in MD simulations on different conformations of hBest1. This forcefield models multipole moments up to the quadrupole; therefore, it captures induced dipole and anion-π interactions. We show that key biophysical properties of the channel can only be simulated when electronic polarization is included in the molecular models and that Cl- permeation through the neck of the pore is achieved through hydrophobic solvation concomitant with partial ion dehydration. Furthermore, we demonstrate how such polarizable simulations can help determine the identity of ion-like densities within high-resolution cryo-EM structures and that neglecting polarization places Cl- at positions that do not correspond with their experimentally resolved location. Overall, our results demonstrate the importance of including electronic polarization in realistic and physically accurate models of biological systems, especially channels and pores that selectively permeate anions.
Keywords: Bestrophin channels; Induced polarization; anion permeation; anion-π; chloride selectivity; molecular dynamics.
Conflict of interest statement
Declaration of interests J.C. is an employee of IBM Research.
Figures


Similar articles
-
Electronic Polarizability Tunes the Function of the Human Bestrophin 1 Cl- Channel.J Chem Theory Comput. 2025 Jan 28;21(2):933-942. doi: 10.1021/acs.jctc.4c01039. Epub 2025 Jan 3. J Chem Theory Comput. 2025. PMID: 39754290 Free PMC article.
-
Influence of effective polarization on ion and water interactions within a biomimetic nanopore.Biophys J. 2022 Jun 7;121(11):2014-2026. doi: 10.1016/j.bpj.2022.05.006. Epub 2022 May 7. Biophys J. 2022. PMID: 35527400 Free PMC article.
-
Cryo-EM structures of mouse bestrophin 1 channel in closed and partially open conformations.Mol Cells. 2025 May;48(5):100208. doi: 10.1016/j.mocell.2025.100208. Epub 2025 Mar 3. Mol Cells. 2025. PMID: 40043778 Free PMC article.
-
Emerging issues of connexin channels: biophysics fills the gap.Q Rev Biophys. 2001 Aug;34(3):325-472. doi: 10.1017/s0033583501003705. Q Rev Biophys. 2001. PMID: 11838236 Review.
-
Mechanisms of cellular synchronization in the vascular wall. Mechanisms of vasomotion.Dan Med Bull. 2010 Oct;57(10):B4191. Dan Med Bull. 2010. PMID: 21040688 Review.
References
-
- Hille B. Ionic Channels of Excitable Membranes, 3rd ed.; Sinauer Associates Inc, 2001.
-
- Ashcroft F. Ion Channels and Disease; Academic Press, 2000.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
