This is a preprint.
Genome folding principles revealed in condensin-depleted mitotic chromosomes
- PMID: 38014261
- PMCID: PMC10680603
- DOI: 10.1101/2023.11.09.566494
Genome folding principles revealed in condensin-depleted mitotic chromosomes
Update in
-
Genome folding principles uncovered in condensin-depleted mitotic chromosomes.Nat Genet. 2024 Jun;56(6):1213-1224. doi: 10.1038/s41588-024-01759-x. Epub 2024 May 27. Nat Genet. 2024. PMID: 38802567
Abstract
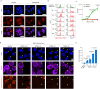
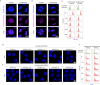
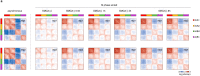
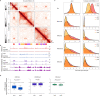
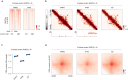
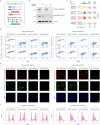
During mitosis, condensin activity interferes with interphase chromatin structures. Here, we generated condensin-free mitotic chromosomes to investigate genome folding principles. Co-depletion of condensin I and II, but neither alone, triggered mitotic chromosome compartmentalization in ways that differ from interphase. Two distinct euchromatic compartments, indistinguishable in interphase, rapidly emerged upon condensin loss with different interaction preferences and dependence on H3K27ac. Constitutive heterochromatin gradually self-aggregated and co-compartmentalized with the facultative heterochromatin, contrasting with their separation during interphase. While topologically associating domains (TADs) and CTCF/cohesin mediated structural loops remained undetectable, cis-regulatory element contacts became apparent, providing an explanation for their quick re-establishment during mitotic exit. HP1 proteins, which are thought to partition constitutive heterochromatin, were absent from mitotic chromosomes, suggesting, surprisingly, that constitutive heterochromatin can self-aggregate without HP1. Indeed, in cells traversing from M- to G1-phase in the combined absence of HP1α, HP1β and HP1γ, re-established constitutive heterochromatin compartments normally. In sum, "clean-slate" condensing-deficient mitotic chromosomes illuminate mechanisms of genome compartmentalization not revealed in interphase cells.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures




















References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
