This is a preprint.
Targeted deletion of Pf prophages from diverse Pseudomonas aeruginosa isolates impacts quorum sensing and virulence traits
- PMID: 38014273
- PMCID: PMC10680813
- DOI: 10.1101/2023.11.19.567716
Targeted deletion of Pf prophages from diverse Pseudomonas aeruginosa isolates impacts quorum sensing and virulence traits
Update in
-
Targeted deletion of Pf prophages from diverse Pseudomonas aeruginosa isolates has differential impacts on quorum sensing and virulence traits.J Bacteriol. 2024 May 23;206(5):e0040223. doi: 10.1128/jb.00402-23. Epub 2024 Apr 30. J Bacteriol. 2024. PMID: 38687034 Free PMC article.
Abstract
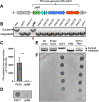
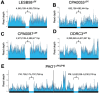
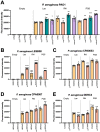
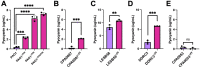
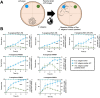
Pseudomonas aeruginosa is an opportunistic bacterial pathogen that commonly causes medical hardware, wound, and respiratory infections. Temperate filamentous Pf phages that infect P. aeruginosa impact numerous bacterial virulence phenotypes. Most work on Pf phages has focused on strain Pf4 and its host P. aeruginosa PAO1. Expanding from Pf4 and PAO1, this study explores diverse Pf strains infecting P. aeruginosa clinical isolates. We describe a simple technique targeting the Pf lysogeny maintenance gene, pflM (PA0718), that enables the effective elimination of Pf prophages from diverse P. aeruginosa hosts. This study also assesses the effects different Pf phages have on host quorum sensing, biofilm formation, virulence factor production, and virulence. Collectively, this research not only introduces a valuable tool for Pf prophage elimination from diverse P. aeruginosa isolates, but also advances our understanding of the complex relationship between P. aeruginosa and filamentous Pf phages.
Figures

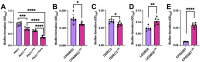
References
-
- Knezevic P, Voet M, Lavigne R. 2015. Prevalence of Pf1-like (pro)phage genetic elements among Pseudomonas aeruginosa isolates. Virology 483:64–71. - PubMed
-
- Secor PR, Burgener EB, Kinnersley M, Jennings LK, Roman-Cruz V, Popescu M, Van Belleghem JD, Haddock N, Copeland C, Michaels LA, de Vries CR, Chen Q, Pourtois J, Wheeler TJ, Milla CE, Bollyky PL. 2020. Pf Bacteriophage and Their Impact on Pseudomonas Virulence, Mammalian Immunity, and Chronic Infections. Front Immunol 11:244. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
